Search Count: 25
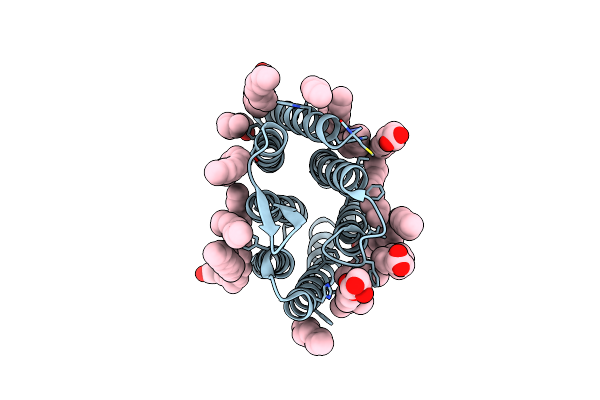 |
Crystal Structure Of Synechocystis Halorhodopsin (Syhr), Cl-Pumping Mode, Ground State
Organism: Synechocystis sp. pcc 7509
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2022-11-09 Classification: MEMBRANE PROTEIN Ligands: CL, LFA, OLC, OLA |
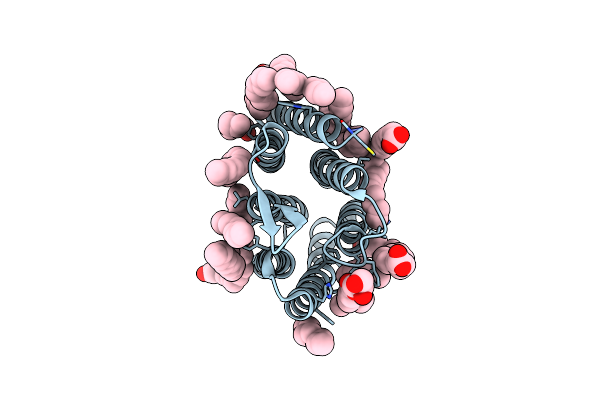 |
Crystal Structure Of Synechocystis Halorhodopsin (Syhr), Cl-Pumping Mode, K State
Organism: Synechocystis sp. pcc 7509
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-11-09 Classification: MEMBRANE PROTEIN Ligands: CL, LFA, OLC, OLA |
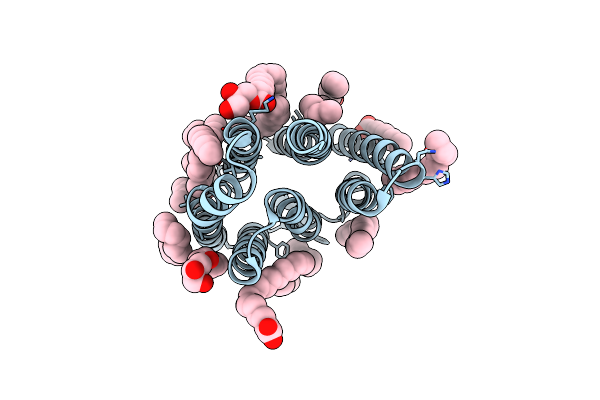 |
Crystal Structure Of Synechocystis Halorhodopsin (Syhr), Cl-Pumping Mode, O State
Organism: Synechocystis sp. pcc 7509
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-09 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC |
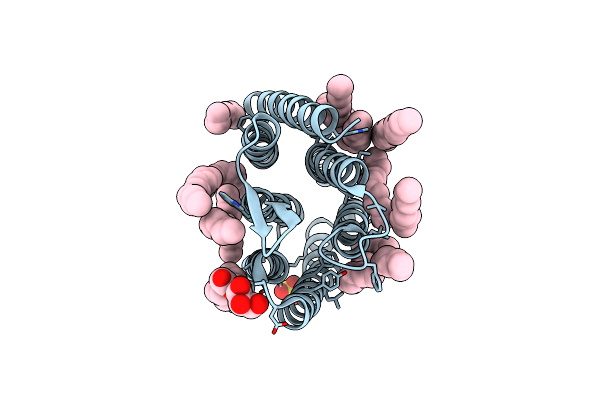 |
Crystal Structure Of Synechocystis Halorhodopsin (Syhr), So4-Bound Form, Ground State
Organism: Synechocystis sp. pcc 7509
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2022-11-09 Classification: MEMBRANE PROTEIN Ligands: CL, SO4, LFA, OLA, GOL |
 |
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC |
 |
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: LFA, 97N |
 |
Crystal Structure Of The D116N Mutant Of The Light-Driven Sodium Pump Kr2 In The Monomeric Form, Ph 4.6
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The D116N Mutant Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The Steady-State-Smx Activated State Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form At Room Temperature, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, NA, RET, GOL, OLA |
 |
Crystal Structure Of The H30A Mutant Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, NA, BOG, RET |
 |
Crystal Structure Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form At Room Temperature, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN |
 |
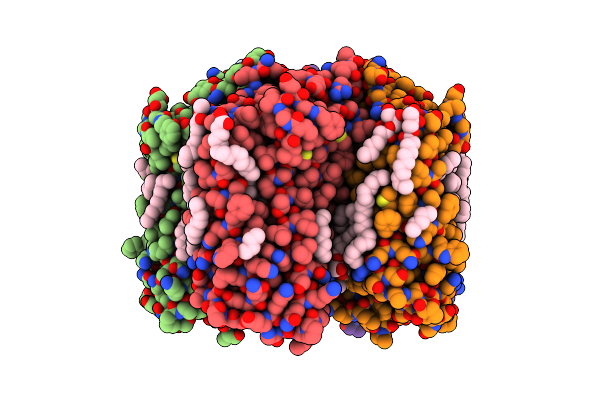
Crystal Structure Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, BOG, RET, NA |
 |
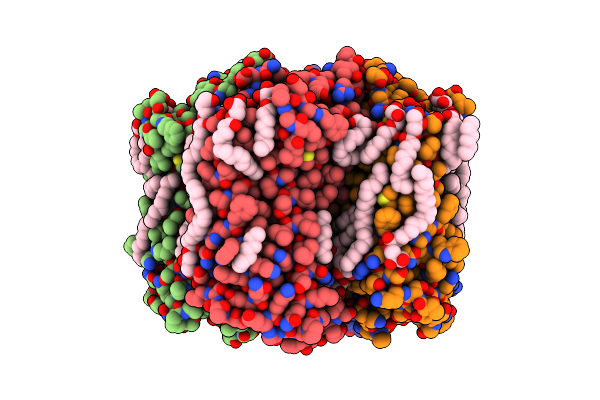
Crystal Structure Of The Steady-State Activated State Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form At Room Temperature, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, NA, RET, GOL, OLA |
 |
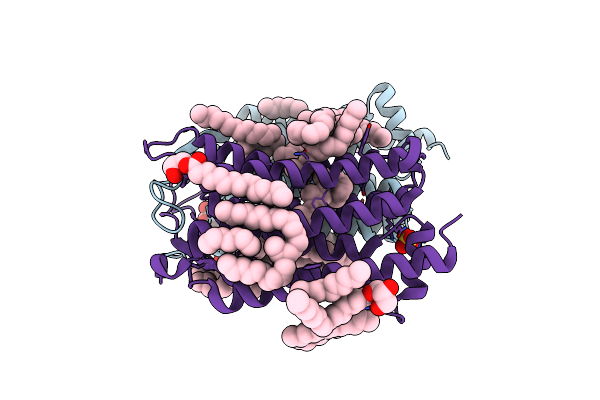
Crystal Structure Of The O-State Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-02-12 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, NA, BOG, OLA, GOL |
 |
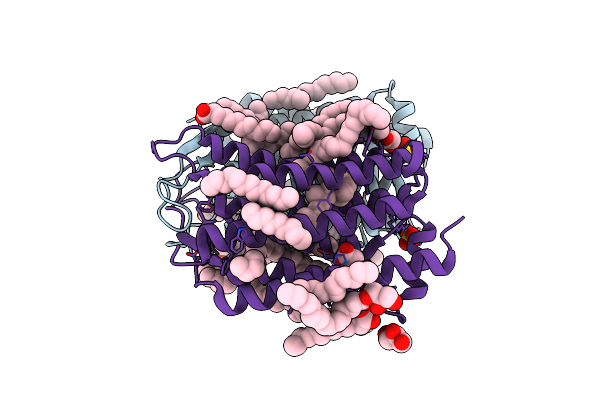
Organism: Actinobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-11 Classification: MEMBRANE PROTEIN Ligands: LFA, OLC, SO4, RET, GOL, OLA |
 |
Organism: Actinobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-11 Classification: MEMBRANE PROTEIN Ligands: ACT, OLA, GOL, OLC, LFA, SO4, RET |
 |
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-11-06 Classification: MEMBRANE PROTEIN Ligands: LFA, RET, OLB |
 |
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-10-04 Classification: HYDROLASE Ligands: ZN, PO4, EDO |
 |
E. Coli Microcin-Processing Metalloprotease Tldd/E With Drvy Angiotensin Fragment Bound
Organism: escherichia coli k-12, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2017-10-04 Classification: HYDROLASE Ligands: ZN, MES, EDO, NA |

