Search Count: 21
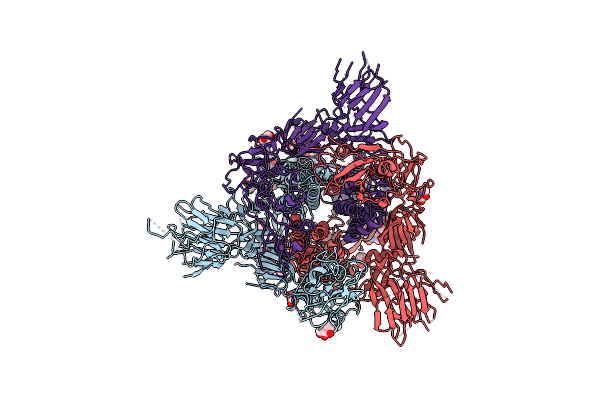 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: VIRAL PROTEIN Ligands: NAG |
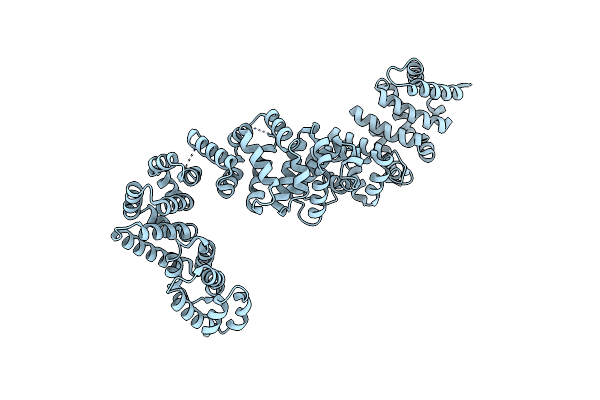 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-06-05 Classification: CHAPERONE |
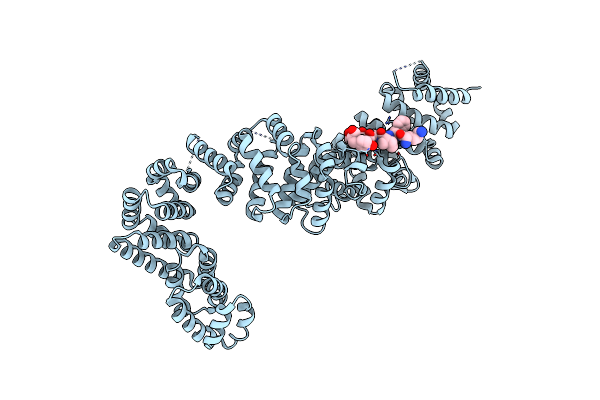 |
Organism: Kluyveromyces lactis, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-06-05 Classification: CHAPERONE |
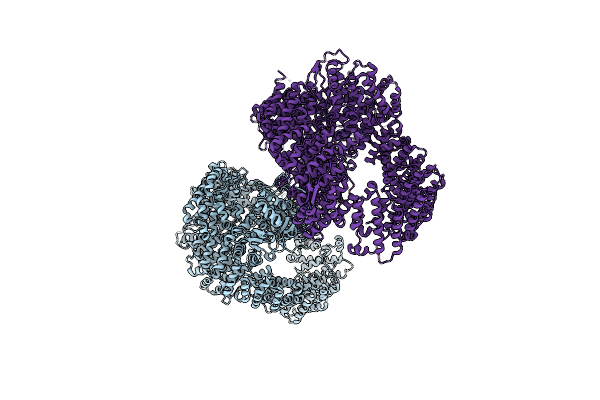 |
Organism: Nematocida sp. ertm5
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2021-07-28 Classification: LIGASE |
 |
Cryo-Em Structure Of Human Ace2 Receptor Bound To Protein Encoded By Vaccine Candidate Bnt162B1
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: VIRAL PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-07-01 Classification: SIGNALING PROTEIN Ligands: ATP, MG, ZN |
 |
Mouse Rnf213 Mutant R4753K Modeling The Moyamoya-Disease-Related Human Variant R4810K
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-07-01 Classification: SIGNALING PROTEIN Ligands: ATP, MG, ZN |
 |
Organism: Starmerella magnoliae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-08 Classification: OXIDOREDUCTASE |
 |
Alcohol Dehydrogenase From Candida Magnoliae Dsmz 70638 (Adha): Complex With Nadp+
Organism: Starmerella magnoliae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-04-08 Classification: OXIDOREDUCTASE Ligands: NAP, IPA, GOL |
 |
Alcohol Dehydrogenase From Candida Magnoliae Dsmz 70638 (Adha): Thermostable 10Fold Mutant
Organism: Starmerella magnoliae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-04-08 Classification: OXIDOREDUCTASE |
 |
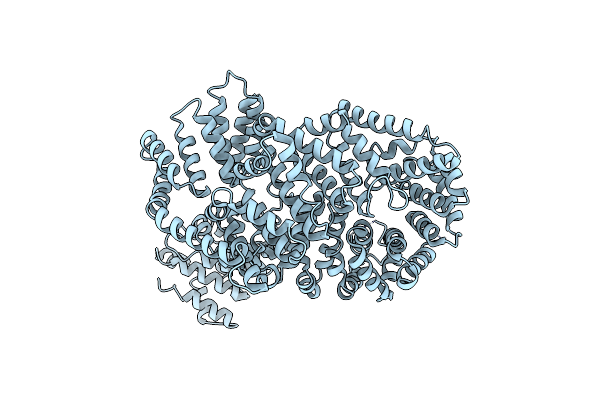
Molecular Features Of The Unc-45 Chaperone Critical For Binding And Folding Muscle Myosin
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2019-10-30 Classification: MOTOR PROTEIN Ligands: ADP, GOL, BU1, PGO, HEZ, P15 |
 |
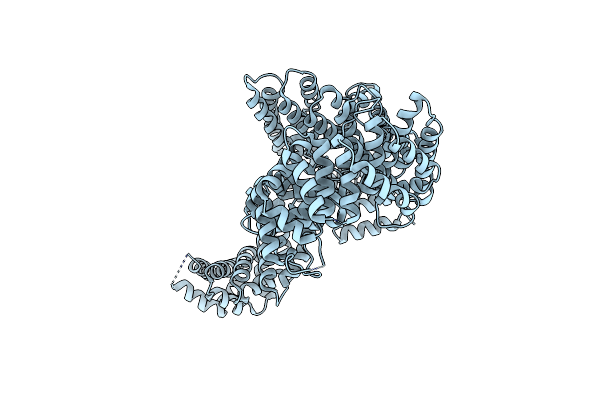
Molecular Features Of The Unc-45 Chaperone Critical For Binding And Folding Muscle Myosin
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2019-10-30 Classification: CHAPERONE |
 |
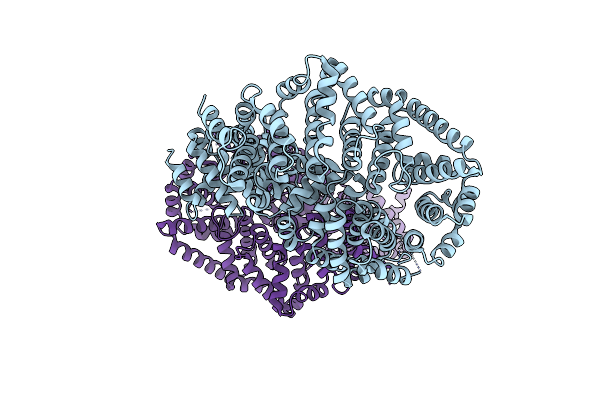
Molecular Features Of The Unc-45 Chaperone Critical For Binding And Folding Muscle Myosin
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2019-10-30 Classification: CHAPERONE |
 |
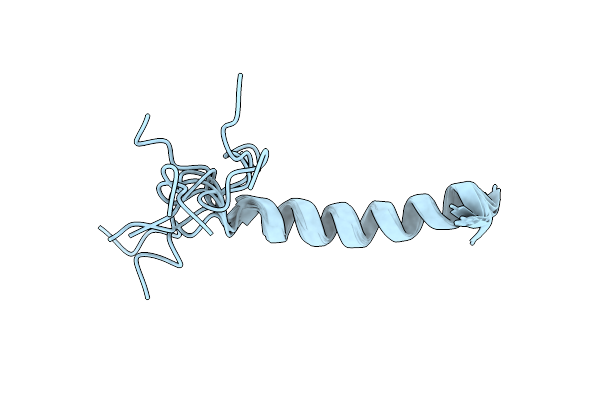
Molecular Features Of The Unc-45 Chaperone Critical For Binding And Folding Muscle Myosin
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2019-10-30 Classification: CHAPERONE |
 |
Organism: Helicobacter pylori
Method: SOLUTION NMR Release Date: 2019-09-11 Classification: ANTITOXIN |
 |
Organism: Helicobacter pylori ss1
Method: SOLUTION NMR Release Date: 2018-05-23 Classification: TOXIN |
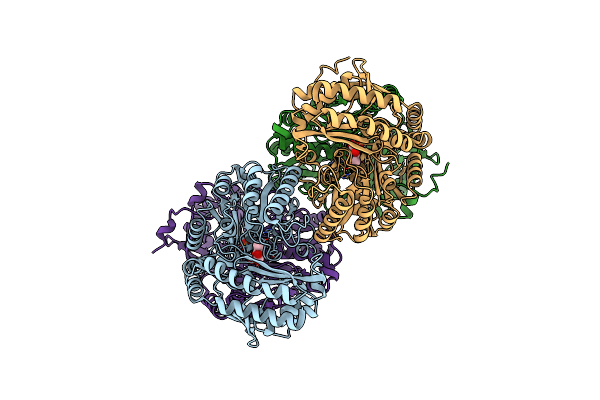 |
Amine Transaminase Crystal Structure From An Uncultivated Pseudomonas Species In The Plp-Bound (Internal Aldimine) Form
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-03-22 Classification: TRANSFERASE Ligands: PLP |
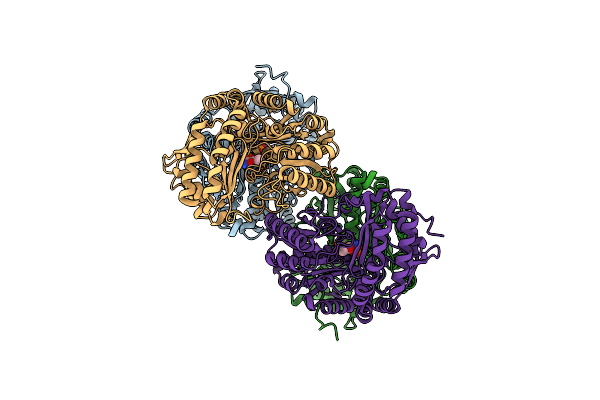 |
Amine Transaminase Crystal Structure From An Uncultivated Pseudomonas Species In The Pmp-Bound Form
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2017-03-22 Classification: TRANSFERASE Ligands: PMP |
 |
Structure Ensemble Backbone And Side-Chain 1H, 13C, And 15N Chemical Shift Assignments, 1H-15N Rdcs And 1H-1H Noe Restraints For Protein K7 From The Vaccinia Virus
Organism: Vaccinia virus
Method: SOLUTION NMR Release Date: 2008-10-28 Classification: VIRAL PROTEIN |

