Search Count: 26
 |
Organism: Clostridium tetanomorphum
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: FLAVOPROTEIN Ligands: FE, SF4, FMN, RBF |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.29 Å Release Date: 2021-06-09 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: DG3 |
 |
Crystal Structure Of The Hiv Capsid Hexamer Bound To The Small Molecule Long-Acting Inhibitor, Gs-6207
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-01 Classification: VIRAL PROTEIN Ligands: QNG |
 |
Discovery Of A Potent Cyclophilin Inhibitor (Compound 3) Based On Structural Simplification Of Sanglifehrin A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-01-25 Classification: isomerase/isomerase inhibitor Ligands: 7HG |
 |
Discovery Of A Potent Cyclophilin Inhibitor (Compound 5) Based On Structural Simplification Of Sanglifehrin A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-25 Classification: isomerase/isomerase inhibitor Ligands: 78E |
 |
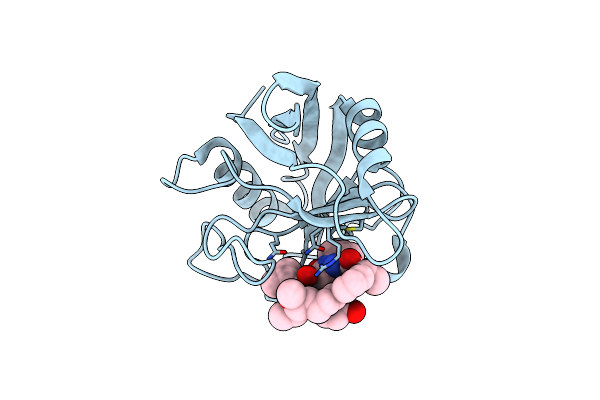
Discovery Of A Potent Cyclophilin Inhibitor (Compound 6) Based On Structural Simplification Of Sanglifehrin A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-01-25 Classification: isomerase/isomerase inhibitor Ligands: 78R |
 |
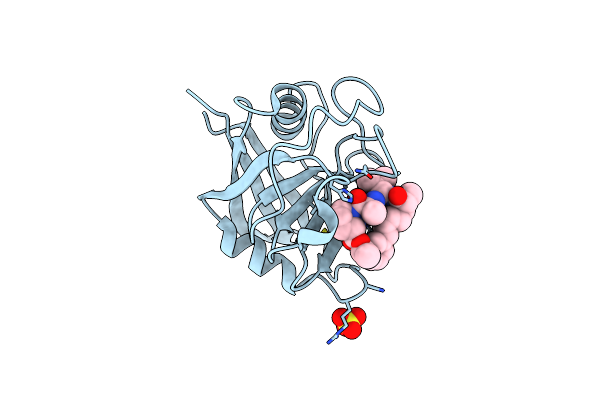
Discovery Of A Potent Cyclophilin Inhibitor (Compound 7) Based On Structural Simplification Of Sanglifehrin A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2017-01-25 Classification: isomerase/isomerase inhibitor Ligands: 78X |
 |
Discovery Of A Potent Cyclophilin Inhibitor (Compound 8) Based On Structural Simplification Of Sanglifehrin A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-01-25 Classification: isomerase/isomerase inhibitor Ligands: 838, SO4 |
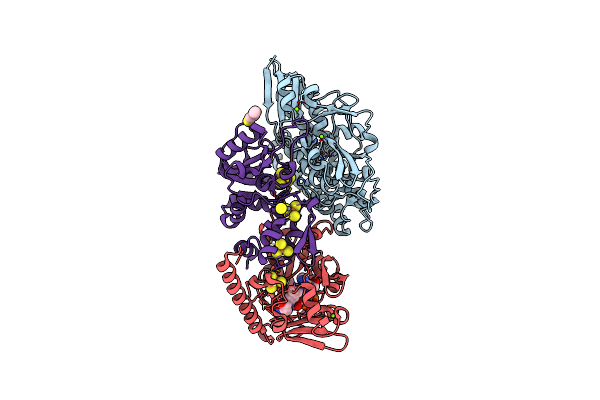 |
The F420-Reducing [Nife]-Hydrogenase Complex From Methanothermobacter Marburgensis, The First X-Ray Structure Of A Group 3 Family Member
Organism: Methanothermobacter marburgensis
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2014-06-11 Classification: OXIDOREDUCTASE Ligands: SF4, DTZ, UNL, MG, NFU, CL, KEN, FAD |
 |
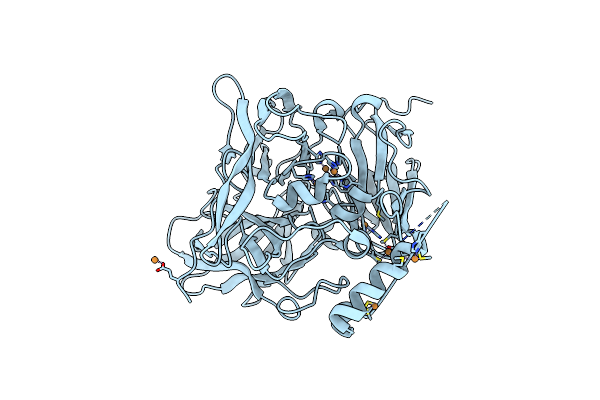
Cryo-Em Structure Of The F420-Reducing Nife-Hydrogenase From A Methanogenic Archaeon With Bound Substrate
Organism: Methanothermobacter marburgensis
Method: ELECTRON MICROSCOPY Resolution:4.00 Å Release Date: 2013-03-06 Classification: OXIDOREDUCTASE Ligands: FCO, NI, FE2, SF4, F42, FAD |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-02-29 Classification: OXIDOREDUCTASE Ligands: CU, CU1, O |
 |
Cueo At 1.1 A Resolution Including Residues In Previously Disordered Region
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2011-09-07 Classification: OXIDOREDUCTASE Ligands: CU, O, EDO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-08-17 Classification: OXIDOREDUCTASE Ligands: CU, SO4, ACT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-17 Classification: OXIDOREDUCTASE Ligands: CU, AG, O |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-17 Classification: OXIDOREDUCTASE |
 |
The Multi-Copper Oxidase Cueo With Six Met To Ser Mutations (M358S,M361S,M362S,M364S,M366S,M368S)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-08-17 Classification: OXIDOREDUCTASE Ligands: CU, C2O |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-08-17 Classification: OXIDOREDUCTASE Ligands: CU1, ACT |
 |
Crystal Structure Of Hiv-1 Rnase H P15 With Engineered E. Coli Loop And Pyrimidinol Carboxylic Acid Inhibitor
Organism: Human immunodeficiency virus type 1 group m subtype b, Escherichia coli (strain k12), Human immunodeficiency virus type 1 group m subtype b (isolate hxb2)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-04-20 Classification: TRANSFERASE, HYDROLASE/INHIBITOR Ligands: MN, SO4, P1Y |
 |
Crystal Structure Of Hiv-1 Rnase H With Engineered E. Coli Loop And N-Hydroxy Quinazolinedione Inhibitor
Organism: Human immunodeficiency virus type 1 group m subtype b, Escherichia coli (strain k12), Human immunodeficiency virus type 1 group m subtype b (isolate hxb2)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2011-04-20 Classification: TRANSFERASE, HYDROLASE/INHIBITOR Ligands: MN, SO4, QID |
 |
Structure Of Hiv-1 Reverse Transcriptase In Complex With An Rnase H Inhibitor And Nevirapine
Organism: Hiv-1 m:b_hxb2r
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2011-04-20 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: NVP, MN, SO4, CL, P4Y |

