Search Count: 52
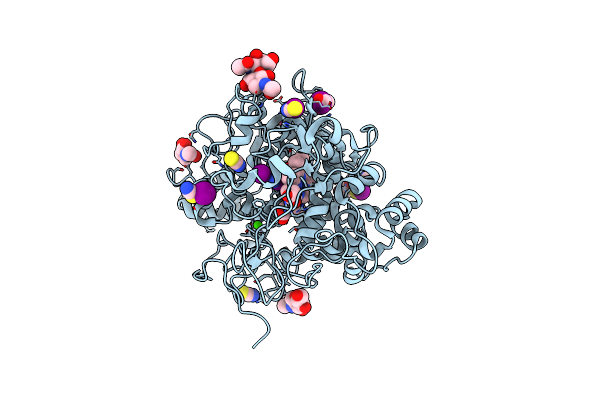 |
Structure Of The Intermediate Of Lactoperoxidase Formed With Thiocynate And Hydrogen Peroxidase At 1.99 A Resolution.
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, SCN, PEO, GOL |
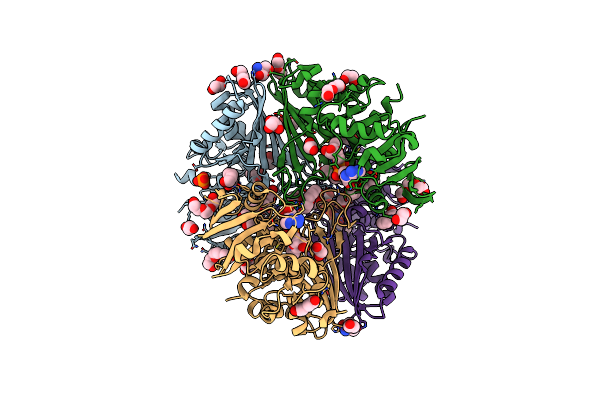 |
Structure Of The Complex Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii With Nicotinamide Adenine Dinucleotide In The Presence Of Poly(Ethylene Glycol) At 2.20 A Resolution
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: NAD, PEG, PG4, EDO, PO4, PGE, TRS, 1PE, GOL |
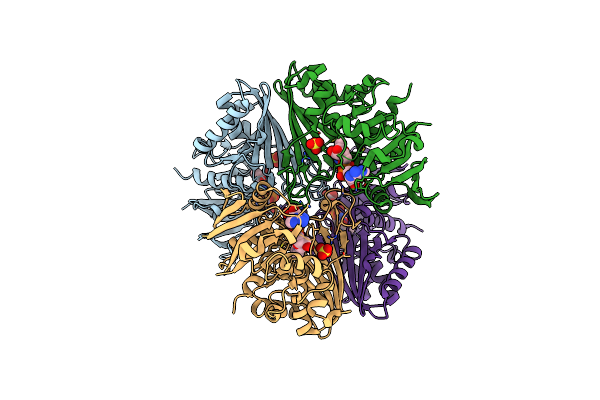 |
Structure Of The Complex Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii With Nicotinamide Adenine Dinucleotide At 2.74 A Resolution.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
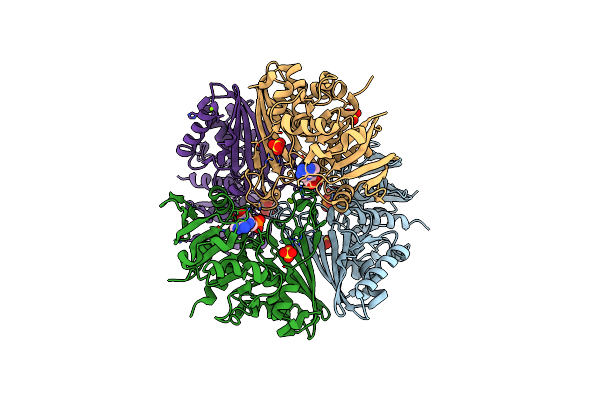 |
Crystal Structure Of The Complex Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii With Adenosine Phosphate At 2.40 A Resolution.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: AMP, SO4, MG |
 |
Crystal Structure Of The Complex Of Glyceraldehyde-3-Phosphate Dehydrogenase Of Type B From Acinetobacter Baumannii With Adenosine Monophosphate At 3.20 A Resolution.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-06-12 Classification: OXIDOREDUCTASE Ligands: AMP, SO4 |
 |
Structure Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii At 3.00 A Resolution
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-06-05 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
 |
Crystal Structure Of Poly(Ethylene Glycol) Stabilized Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii At 2.30 A Resolution
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-06-05 Classification: OXIDOREDUCTASE Ligands: NAD, PEG, PG4, EDO, TRS, GOL, XPE, PGE, SO4, MG |
 |
Crystal Structure Of The Complex Of Lactoperoxidase With Four Inorganic Substrates, Scn, I, Br And Cl
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-13 Classification: OXIDOREDUCTASE Ligands: HEM, CA, IOD, BR, CL, PEG, SCN, NAG |
 |
Crystal Structure Of The Complex Of Phosphopantetheine Adenylyltransferase From Acinetobacter Baumannii With Dephosphocoenzyme-A At 2.19 A Resolution.
Organism: Acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-04-12 Classification: TRANSFERASE Ligands: MG, COD, SO4, CL |
 |
Structure Of The Ternary Complex Of Lactoperoxidase With Substrate Nitric Oxide (No) And Product Nitrite Ion (No2) At 1.98 A Resolution
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: NAG, NO, NO2, NO3, SCN, EDO, IOD, HEM, CA, OSM, NA |
 |
Structure Of The Complex Of Lactoperoxidase With Nitric Oxide Catalytic Product Nitrite At 1.89 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2023-01-11 Classification: OXIDOREDUCTASE Ligands: CA, IOD, SCN, EDO, NO2, HEM, ZN, NAG |
 |
Crystal Structure Of The Complex Of Lactoperoxidase With Nitric Oxide At 2.50A Resolution
Organism: Bos grunniens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-06-29 Classification: OXIDOREDUCTASE Ligands: MH0, CA, CL, NAG, NO, ACT |
 |
Structure Of The Complex Of Camel Peptidoglycan Recognition Protein-Short (Pgrp-S) With Heptanoic Acid At 2.15 A Resolution.
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-06-15 Classification: IMMUNE SYSTEM Ligands: EDO, PEO, CO3, NA, SHV, MPD |
 |
Crystal Structure Of The Ternary Complex Of Peptidoglycan Recognition Protein, Pgrp-S With Hexanoic And Tartaric Acids At 2.07 A Resolution.
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2022-05-11 Classification: IMMUNE SYSTEM Ligands: TLA, EDO, ACT, CL, 6NA, GOL |
 |
Crystal Structure Of The Ternary Complex Of Peptidoglycan Recognition Protein, Pgrp-S With Hexanoic And Tartaric Acids At 2.28 A Resolution.
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-05-11 Classification: IMMUNE SYSTEM Ligands: TLA, EDO, CL, 6NA, ACT, GOL |
 |
Crystal Structure Of The Ternary Complex Of Peptidoglycan Recognition Protein, Pgrp-S With Hexanoic And Tartaric Acids At 2.67 A Resolution.
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2022-05-11 Classification: IMMUNE SYSTEM Ligands: EDO, TLA, CL, ACT, 6NA, GOL |
 |
Crystal Structure Of Adenosine Triphosphate Phosphoribosyltransferase (Hisg) From Acinetobacter Baumannii At 2.975 A Resolution
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: FMT, GOL |
 |
Crystal Structure Of Adenosine Triphosphate Phosphoribosyltransferase (Hisg) From Acinetobacter Baumannii At 3.13 A Resolution
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:3.13 Å Release Date: 2022-01-19 Classification: TRANSFERASE |
 |
Crystal Structure Of Adenosine Triphosphate Phosphoribosyltransferase (Hisg) From Acinetobacter Baumannii At 3.15 A Resolution
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2022-01-19 Classification: TRANSFERASE Ligands: ACT |
 |
Crystal Structure Of Pepsin Cleaved Lactoferrin C-Lobe At 2.28 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2021-08-04 Classification: HYDROLASE Ligands: FE, CO3, NAG |

