Search Count: 25
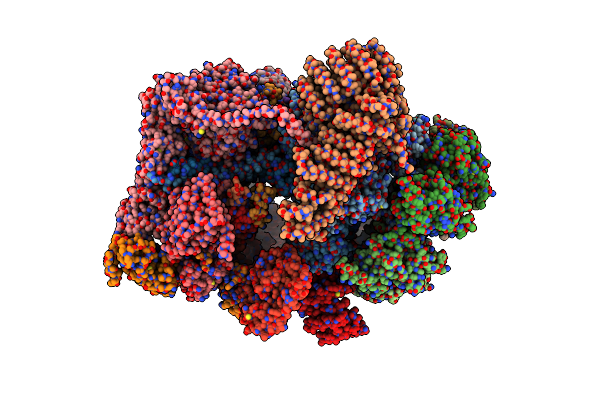 |
Cryo-Em Structure Of Human 26S Rp (Ed State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Four Ub.
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
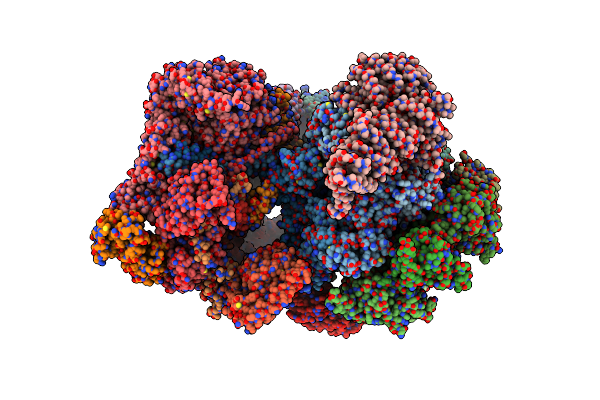 |
Cryo-Em Structure Of Human 26S Rp (Eb State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Four Ub.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
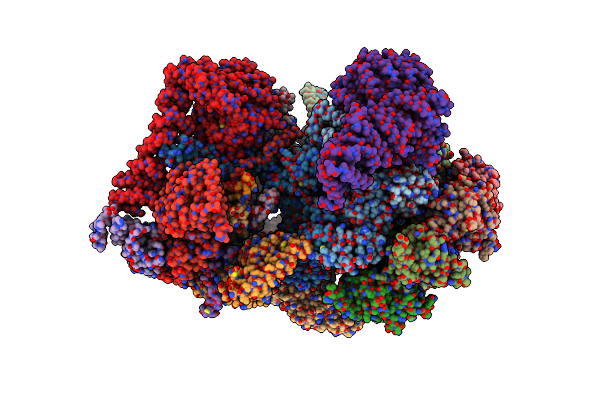 |
Cryo-Em Structure Of Human 26S Proteasomal Rp Subcomplex (Ea State) Without Any Bound Substrate.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
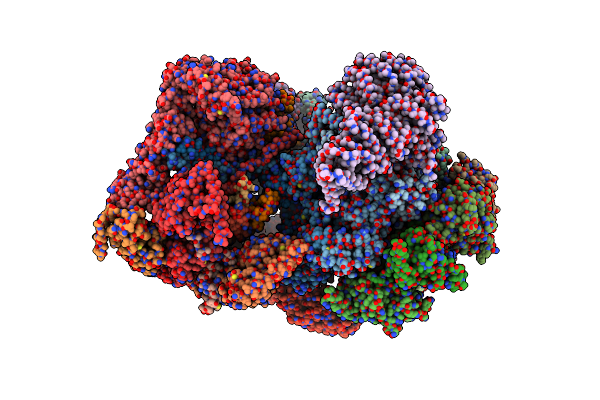 |
Cryo-Em Structure Of Human 26S Proteasomal Rp Subcomplex (Ea State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Three Ub.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of Gpr119-Gs-Nb35 Complex With Small Molecule Agonist Mbx-2982
Organism: Homo sapiens, Lama glama, Escherichia coli, Oplophorus gracilirostris
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: MEMBRANE PROTEIN/SIGNALING PROTEIN Ligands: 8VP |
 |
Cryoem Structure Of An Inward-Facing Melbst At A Na(+)-Bound And Sugar Low-Affinity Conformation
Organism: Salmonella enterica subsp. enterica serovar typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-02-28 Classification: TRANSPORT PROTEIN |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSFERASE |
 |
Organism: Adeno-associated virus - 1
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-02-13 Classification: HYDROLASE Ligands: FNB |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2019-01-02 Classification: HYDROLASE Ligands: CW5, MG, GOL |
 |
Crystal Structure Of Igpd From Pyrococcus Furiosus In Complex With (R)-C348
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-05 Classification: LYASE Ligands: MN, 5LD |
 |
A. Thaliana Igpd2 In Complex With The Racemate Of The Triazole-Phosphonate Inhibitor, C348
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2016-10-05 Classification: LYASE Ligands: MN, 5DL, 5LD, EDO, TRS, CL |
 |
A. Thaliana Igpd2 In Complex With The Triazole-Phosphonate Inhibitor, (S)-C348, To 1.1A Resolution
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2016-10-05 Classification: LYASE Ligands: MN, 5DL, TRS, EDO |
 |
A. Thaliana Igpd2 In Complex With The Triazole-Phosphonate Inhibitor, (R)-C348, To 1.36A Resolution
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-10-05 Classification: LYASE Ligands: MN, 5LD, EDO, TRS, CL |
 |
Crystal Structure Of Igpd From Pyrococcus Furiosus In Complex With (S)-C348
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2016-09-28 Classification: LYASE Ligands: MN, 5DL |
 |
The Structure Of Wt A. Thaliana Igpd2 In Complex With Mn2+ And Formate At 1.3A Resolution
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2015-06-24 Classification: LYASE Ligands: MN, FMT, TRS, CL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2015-06-24 Classification: LYASE Ligands: MN, PO4, NA, EDO |
 |
The Structure Of Wt A. Thaliana Igpd2 In Complex With Mn2+ And 1,2,4-Triazole At 1.3 A Resolution
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-09-24 Classification: LYASE Ligands: MN, TRI, TRS, CL, EDO |
 |
The Structure Of Wt A. Thaliana Igpd2 In Complex With Mn2+, Imidazole, And Sulfate At 1.5 A Resolution
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-09-24 Classification: LYASE Ligands: MN, IMD, SO4, EDO |
 |
The Form A Structure Of An E21Q Catalytic Mutant Of A. Thaliana Igpd2 In Complex With Mn2+ And A Mixture Of Its Substrate, 2R3S-Igp, And An Inhibitor, 2S3S-Igp, To 1.12 A Resolution
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2014-09-24 Classification: LYASE/LYASE INHIBITOR Ligands: MN, IG2, IYP, EDO |

