Search Count: 1,621
 |
Structure Of The Transaminase Phnw From Vibrio Vulnificus In Complex With Plp
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE |
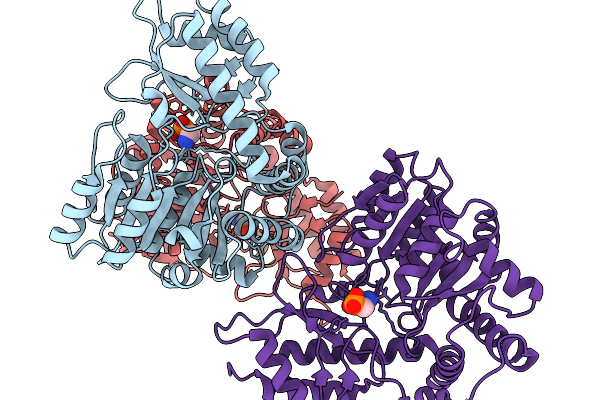 |
Structure Of The Transaminase Phnw From Vibrio Vulnificus In Complex With Plp And Aep
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: P7I |
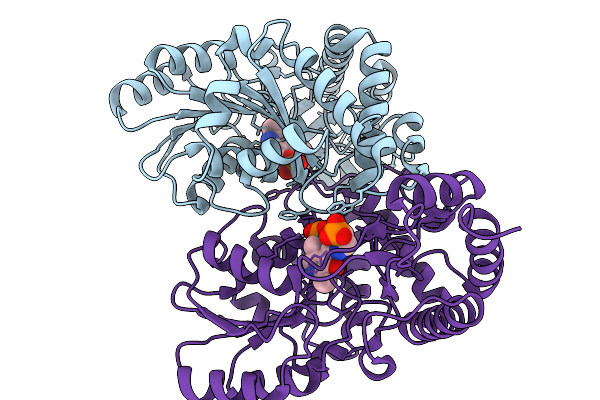 |
Structure Of The K193M Mutant Of Transaminase Phnw From Vibrio Vulnificus In Complex With Plp And Aep
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: PLP, P7I, CL |
 |
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN Ligands: A1AFF |
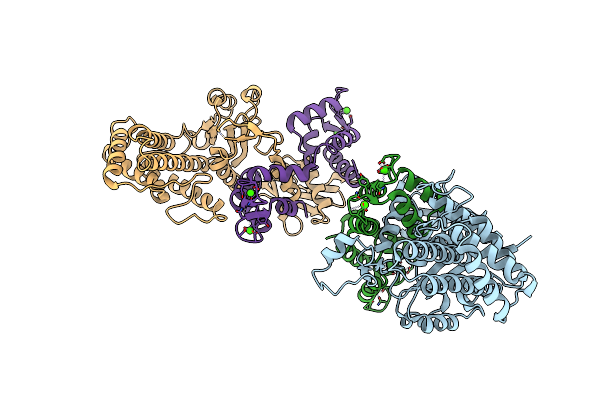 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: CA |
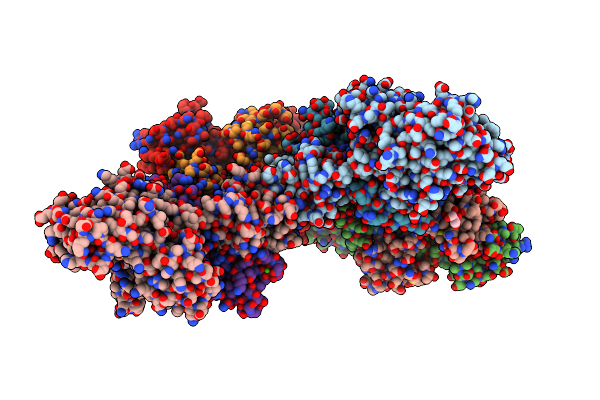 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin And A Nicotinamide Adenine Dinucleotide (Nad+)
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: NAD, CA, GOL |
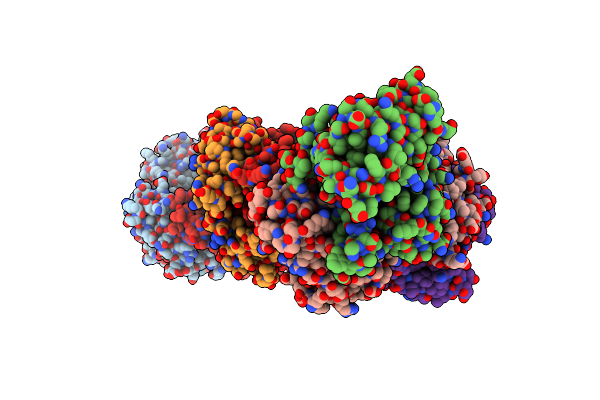 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Free Calmodulin
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: MG |
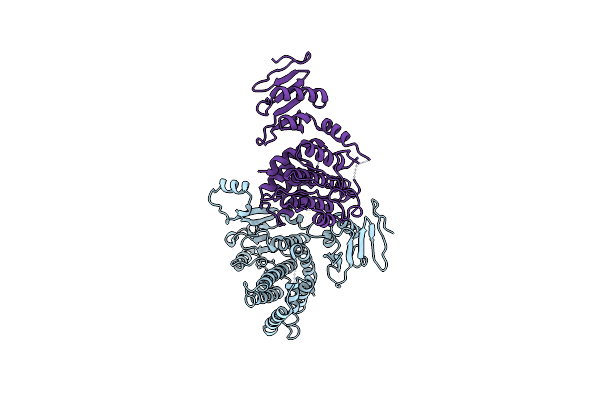 |
Crystal Structure Of The Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) From Vibrio Vulnificus
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2024-07-10 Classification: TOXIN |
 |
Structure Of Vibrio Vulnificus Martx Cysteine Protease Domain Lacking Beta-Flap
Organism: Vibrio vulnificus mo6-24/o
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-10 Classification: TOXIN |
 |
Organism: Vibrio vulnificus mo6-24/o
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-07-10 Classification: TOXIN |
 |
X-Ray Structure Of The Aminotransferase From Vibrio Vulnificus Responsible For The Biosynthesis Of 2,3-Diacetamido-4-Amino-2,3,4-Trideoxy-Arabinose In The Presence Of Its Internal Aldimine
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: EDO, CL |
 |
X-Ray Structure Of The Aminotransferase From Vibrio Vulnificus Responsible For The Biosynthesis Of 2,3-Diacetamido-4-Amino-2,3,4-Trideoxy-Arabinose In The Presence Of Its External Aldimine With 2,3-Diacetamido-4-Amino-2,3,4-Trideoxy-L-Arabinose
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: A1APE, MES, EDO, PGE |
 |
Crystal Structure Of The Open Unbound Catalytically Inactive Makes Caterpillars Floppy-Like (Mcf) Effector From Vibrio Vulnificus Cmcp6
Organism: Vibrio vulnificus cmcp6
Method: X-RAY DIFFRACTION Release Date: 2024-06-05 Classification: TOXIN Ligands: SO4, CL |
 |
Organism: Vibrio vulnificus yj016
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-02-07 Classification: CYTOSOLIC PROTEIN Ligands: TPP, EDO, MG, NI |
 |
Organism: Vibrio vulnificus cmcp6
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-07-05 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Siderophore Binding Protein Vatd From Vibrio Vulnificus M2799 Complexed With Desferal
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-12-07 Classification: METAL BINDING PROTEIN Ligands: KTY, FE, MG |
 |
Structure Of Oxygen-Insensitive Nad(P)H-Dependent Nitroreductase Nfsb_Vv F70A/F108Y (Ntr 2.0) In Complex With Fmn At 1.85 Angstroms Resolution
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: FMN, ACT, P4K |
 |
Organism: Vibrio vulnificus
Method: ELECTRON MICROSCOPY Release Date: 2022-04-13 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of The Intact Mtase From Vibrio Vulnificus Yj016 In Complex With The Dna-Mimicking Ocr Protein And The S-Adenosyl-L-Homocysteine (Sah)
Organism: Vibrio vulnificus (strain yj016), Escherichia phage t7
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-03-23 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-02-16 Classification: TRANSFERASE Ligands: SAH |

