Search Count: 37
All
Selected
 |
Organism: Vibrio proteolyticus nbrc 13287
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-12-29 Classification: TRANSFERASE Ligands: PLP, MES, SO4, GLY |
 |
Organism: Vibrio proteolyticus nbrc 13287
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2021-12-29 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of Aeromonas Proteolytica Aminopeptidase Complexed With 8-Quinolinol
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2012-05-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, HQY, NA, CL, SCN, GOL |
 |
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-11-17 Classification: HYDROLASE Ligands: ZN, TRS, NA |
 |
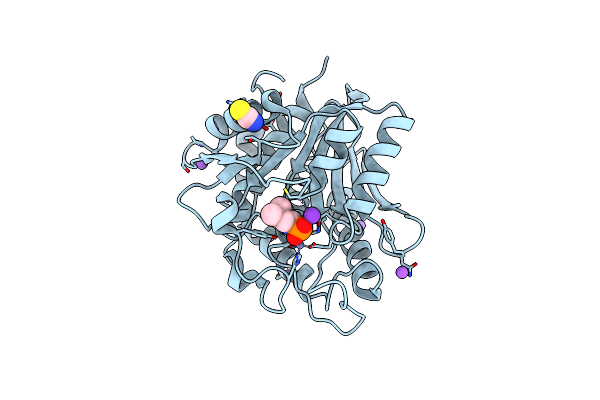
Crystal Structure Of The M180A Mutant Of The Aminopeptidase From Vibrio Proteolyticus
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2007-11-27 Classification: HYDROLASE Ligands: ZN, NA, SCN |
 |
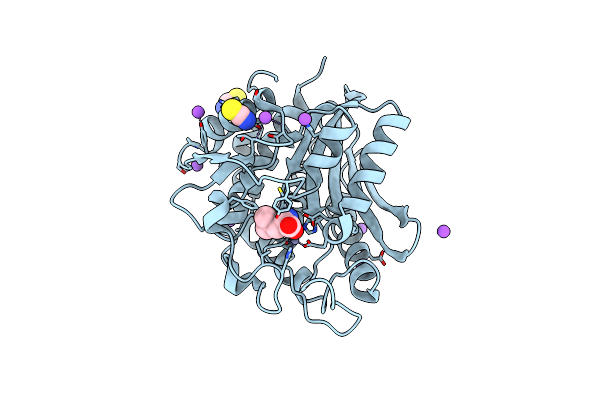
Crystal Structure Of The M180A Mutant Of The Aminopeptidase From Vibrio Proteolyticus In Complex With Leucine Phosphonic Acid
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2007-11-27 Classification: HYDROLASE Ligands: ZN, K, SCN, NA, PLU |
 |
Crystal Structure Of The M180A Mutant Of The Aminopeptidase From Vibrio Proteolyticus In Complex With Leucine
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2007-11-27 Classification: HYDROLASE Ligands: ZN, NA, SCN, LEU |
 |
Crystal Structure Of The D118N Mutant Of The Aminopeptidase From Vibrio Proteolyticus
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2007-11-27 Classification: HYDROLASE Ligands: ZN, NA, SCN, ILE |
 |
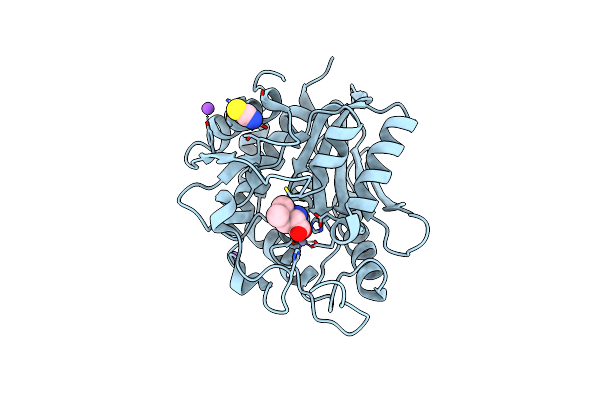
Crystal Structure Of The S228A Mutant Of The Aminopeptidase From Vibrio Proteolyticus
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2007-11-27 Classification: HYDROLASE Ligands: ZN, NA, SCN, VAL |
 |
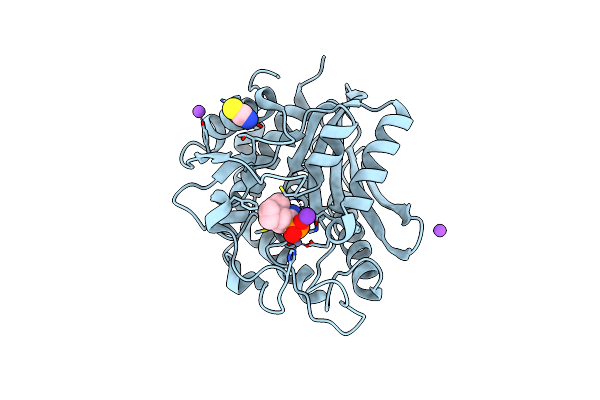
Crystal Structure Of The S228A Mutant Of The Aminopeptidase From Vibrio Proteolyticus In Complex With Leucine
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-11-27 Classification: HYDROLASE Ligands: ZN, SCN, NA, LEU |
 |
Crystal Structure Of The S228A Mutant Of The Aminopeptidase From Vibrio Proteolyticus In Complex With Leucine Phosphonic Acid
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-11-27 Classification: HYDROLASE Ligands: ZN, K, NA, SCN, PLU, LEU |
 |
Structure Of Vibrio Proteolyticus Aminopeptidase With A Bound Trp Fragment Of Dlwcf
Organism: Synthetic construct, Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-10-30 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of The Aminopeptidase From Vibrio Proteolyticus In Complexation With Leucyl-Leucyl-Leucine.
Organism: Synthetic construct, Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-08-28 Classification: HYDROLASE Ligands: ZN |
 |
X-Ray Crystallographic Characterization Of The Co(Ii)-Substituted Tris-Bound Form Of The Aminopeptidase From Aeromonas Proteolytica
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2007-06-12 Classification: HYDROLASE Ligands: CO, TRS |
 |
Crystal Structure Of The Aminopeptidase Of Aeromonas Proteolytica At Ph 4.7
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2006-07-25 Classification: HYDROLASE Ligands: ZN, NA |
 |
Functional Glutamate 151 To Histidine Mutant Of The Aminopeptidase From Aeromonas Proteolytica.
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-10-04 Classification: HYDROLASE Ligands: ZN, NA |
 |
Crystal Structure Of Aeromonas Proteolytica Aminopeptidase In Complex With Bestatin
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-09-27 Classification: HYDROLASE Ligands: ZN, BES |
 |
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-07-20 Classification: HYDROLASE Ligands: ZN, BES |
 |
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-06-22 Classification: TRANSFERASE Ligands: CA |
 |
Crystal Structure Of Vibrio Proteolyticus Chitobiose Phosphorylase In Complex With Glcnac
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-06-22 Classification: TRANSFERASE Ligands: NDG, NAG, CA, CL |

