Search Count: 14
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: OXIDOREDUCTASE Ligands: FES, 9YF, PLM, HEM, CDL, A1IRE, HEC, MQ9, 9Y0, CU, HEA, CA, MQ7 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structural Insight Of Flavonoids Binding To Cag Repeat Rna That Causes Huntington'S Disease (Hd) And Spinocerebellar Ataxia (Scas)
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-08-08 Classification: RNA Ligands: MYC |
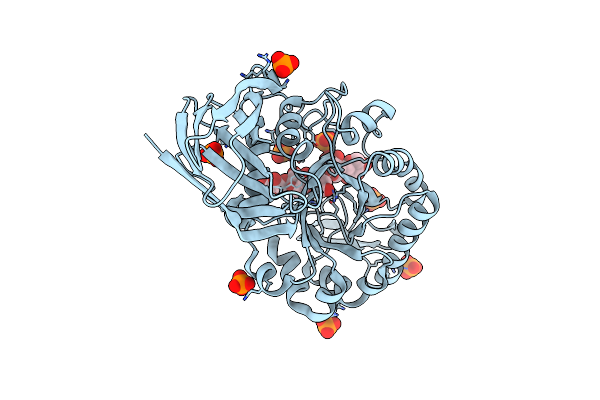 |
High Resolution Structure Of The Thermostable Glucuronoxylan Endo-Beta-1, 4-Xylanase, Ctxyn30A, From Clostridium Thermocellum With Two Xylobiose Units Bound
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-19 Classification: HYDROLASE Ligands: PO4, PEG |
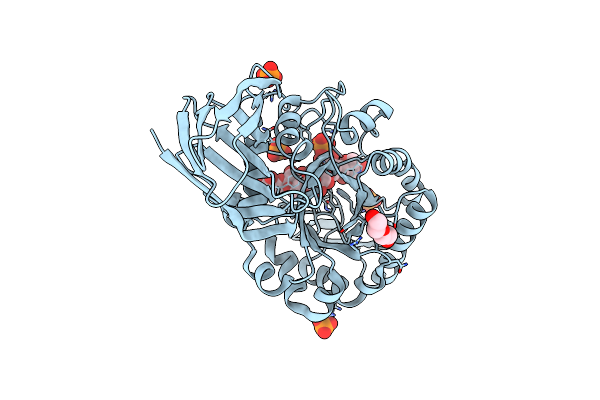 |
Determining The Specificities Of The Catalytic Site From The Very High Resolution Structure Of The Thermostable Glucuronoxylan Endo-Beta-1, 4-Xylanase, Ctxyn30A, From Clostridium Thermocellum With A Xylotetraose Bound
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2016-10-19 Classification: HYDROLASE Ligands: PO4, PEG |
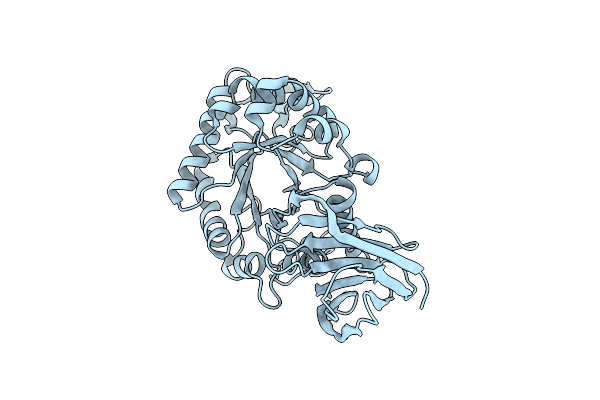 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum At 1.77 A Resolution
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2015-06-24 Classification: HYDROLASE |
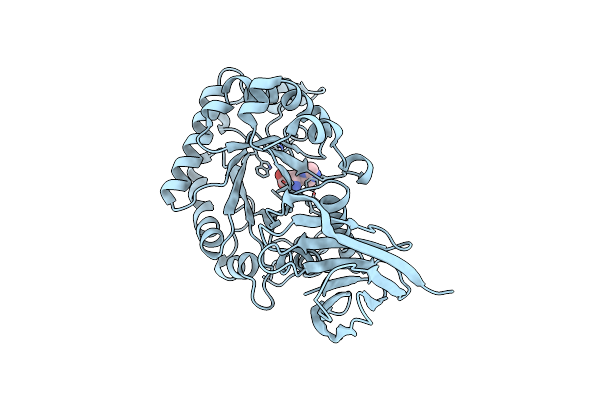 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: HIS |
 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum At 1.68 A Resolution
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: SO4, EPE |
 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum At 1.30 A Resolution
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: TAR, TLA, GLC |
 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum At 1.25 A Resolution
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: GOL, TRS, THR |
 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum At 1.28 A Resolution
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: GOL, TRS, PEG, PGO |
 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum
Organism: Clostridium thermocellum, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-01-21 Classification: HYDROLASE Ligands: MLA |
 |
Structure Of Zinc Induced Heterodimer Of Two Calcium Free Isoforms Of Phospholipase A2 From Naja Naja Sagittifera At 2.7A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2005-03-15 Classification: HYDROLASE Ligands: ZN, ACY |

