Search Count: 4
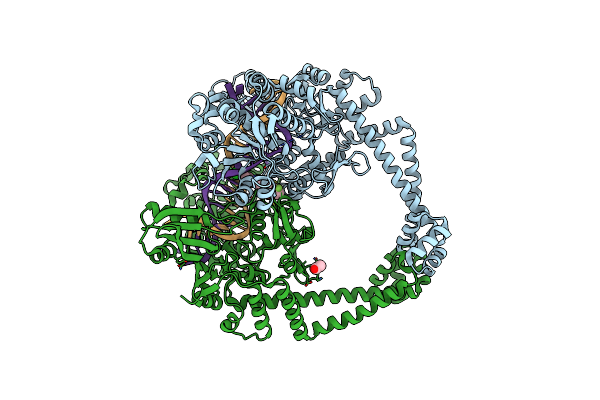 |
Crystal Structure Of S. Aureus Gyrase In Complex With 4-[[1-[(1-Chloro-6,7-Dihydro-5H-Cyclopenta[C]Pyridin-6-Yl)Methyl]Azetidin-3-Yl]Methylamino]-6-Fluorochromen-2-One
Organism: Staphylococcus aureus subsp. aureus n315, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2023-06-28 Classification: ISOMERASE Ligands: MN, CL, ACT, 61I |
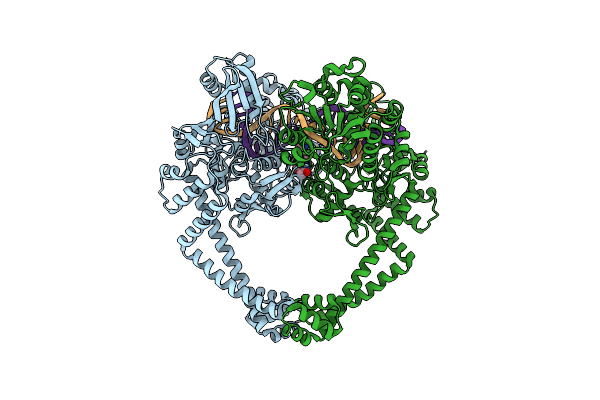 |
Crystal Structure Of S. Aureus Gyrase In Complex With 6-[5-[2-[(4-Chloro-2,3-Dihydro-1H-Inden-2-Yl)Methylamino]Ethyl]-2-Oxo-1,3-Oxazolidin-3-Yl]-4H-Pyrido[3,2-B][1,4]Oxazin-3-One
Organism: Staphylococcus aureus subsp. aureus n315, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2023-06-28 Classification: ISOMERASE Ligands: MN, CL, 6I0 |
 |
The Dna Gyrase B Atp Binding Domain Of Pseudomonas Aeruginosa In Complex With Compound 12X
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-02 Classification: ISOMERASE Ligands: DMS, SO4, NA, EZ6 |
 |
The Dna Gyrase B Atp Binding Domain Of Pseudomonas Aeruginosa In Complex With Compound 12O
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-09-02 Classification: ISOMERASE Ligands: EZ9, SO4, CL |

