Search Count: 138
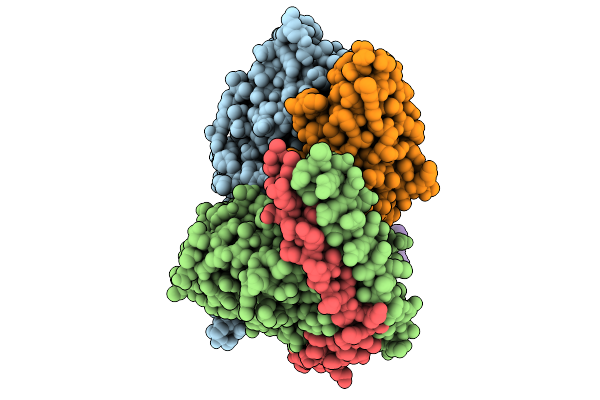 |
Human M4 Muscarinic Acetylcholine Receptor Gi1 Complex (Mini-Gsi Chimera) Bound To The Bitopic Agonist C110
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: SIGNALING PROTEIN Ligands: 5XI |
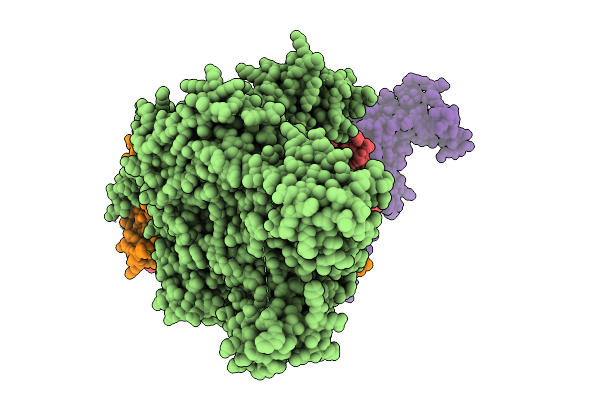 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.34 Å Release Date: 2026-01-21 Classification: CELL CYCLE |
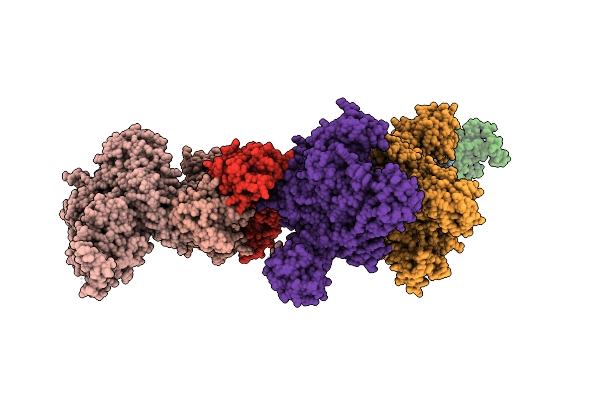 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:5.00 Å Release Date: 2026-01-21 Classification: CELL CYCLE |
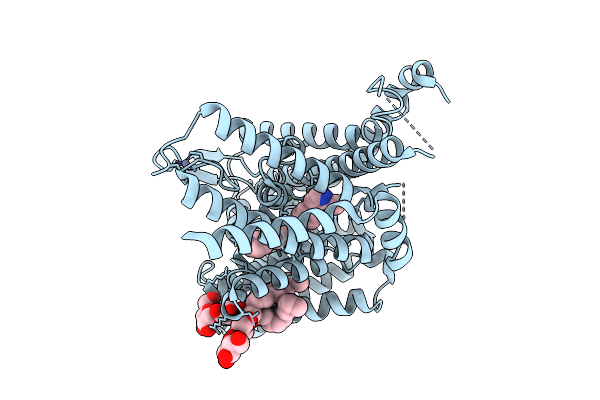 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: A1CC5, AV0, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: ZN, A1CC6 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Resolution:1.95 Å Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Human Delta Opioid Receptor Complex With Mini-Gi And Agonist Dadle And Allosteric Modulator Mips3614
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Resolution:1.94 Å Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: A1CVG |
 |
Human Delta Opioid Receptor Complex With Mini-Gi And Agonist Dadle And Allosteric Modulator Mips3983
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Resolution:2.14 Å Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: A1CU8 |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN |
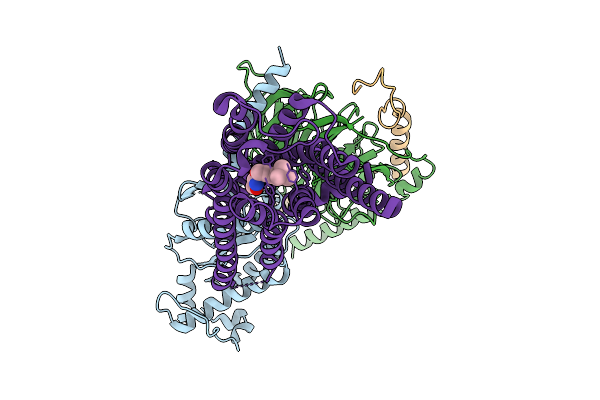 |
Human M5 Muscarinic Acetylcholine Receptor Complex With Mini-Gq And Iperoxo
Organism: Homo sapiens, Rattus norvegicus
Method: ELECTRON MICROSCOPY Resolution:2.79 Å Release Date: 2025-10-08 Classification: MEMBRANE PROTEIN Ligands: IXO |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN Ligands: ADN |
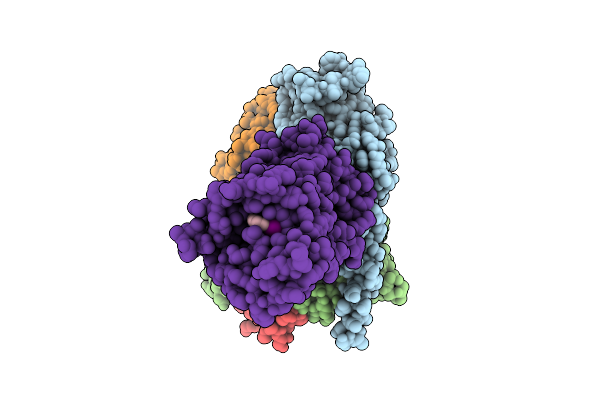 |
Human Adenosine A3 Receptor Gi Complex (Mini-Gsi Chimera) Bound To Piclidenoson (Cf101, Ib-Meca)
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN Ligands: Q8L |
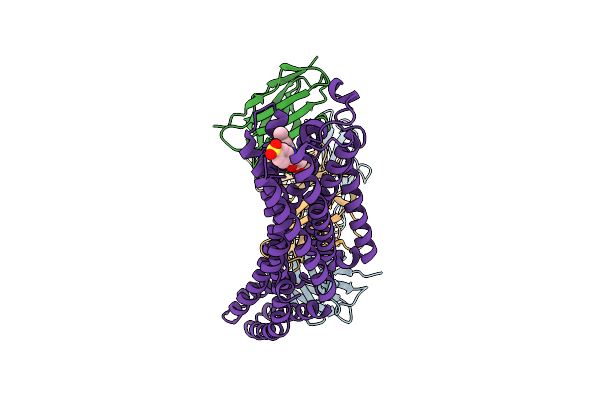 |
Structure Of A Human Adenosine A3 Receptor Complex Bound To The Covalent Antagonist Luf7602
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: SIGNALING PROTEIN Ligands: A1BII |
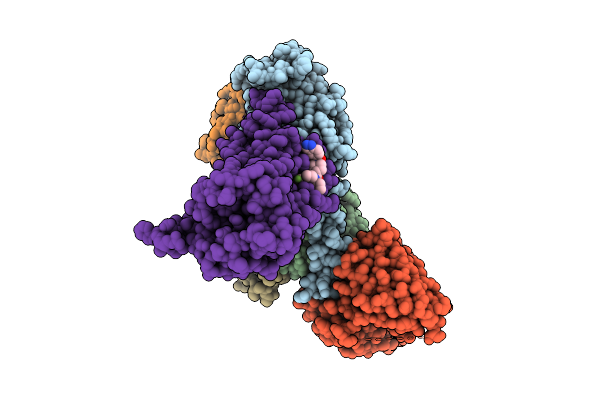 |
Human M5 Muscarinic Acetylcholine Receptor Complex With Mini-Gq, Agonist Acetylcholine And Positive Allosteric Modulator Vu6007678
Organism: Lama glama, Rattus norvegicus, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:2.06 Å Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: A1BKS, ACH |
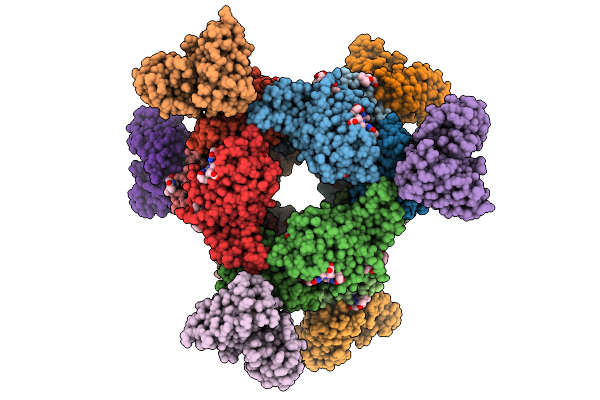 |
Organism: Mus musculus, Neopyropia tenera
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: IMMUNE SYSTEM Ligands: PEB, PUB |
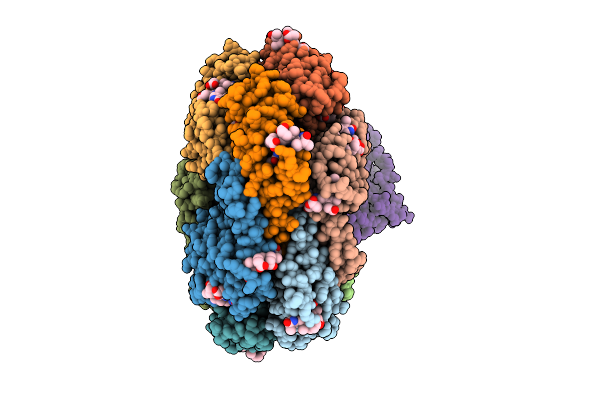 |
Organism: Mus musculus, Ceramium secundatum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: IMMUNE SYSTEM Ligands: PEB, PUB |
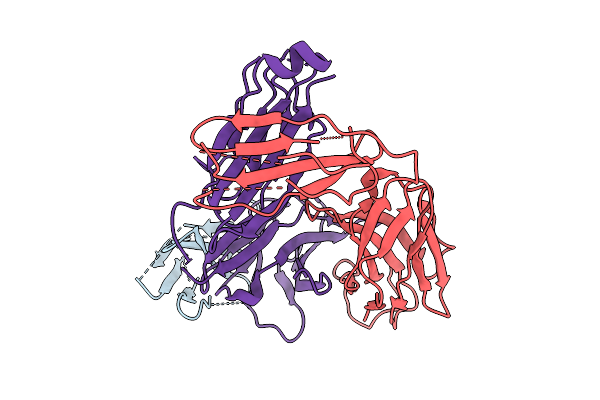 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: IMMUNE SYSTEM |

