Search Count: 23
 |
Crystal Structure Of A Bacterial Pyranose 2-Oxidase In Complex With Mangiferin
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: FAD, HZI, SO4 |
 |
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-07-05 Classification: OXIDOREDUCTASE Ligands: FAD, GLC, SO4, TRS |
 |
Crystal Structure Of A Bacterial Pyranose 2-Oxidase From Pseudoarthrobacter Siccitolerans
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-06-14 Classification: OXIDOREDUCTASE Ligands: SO4, TRS, FAD |
 |
Organism: Mycolicibacterium hassiacum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-01-25 Classification: TRANSFERASE |
 |
Organism: Mycolicibacterium hassiacum
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: MG, NI, GOL |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) - Apo Form
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2019-05-15 Classification: HYDROLASE |
 |
Structure Of Met1 From Mycobacterium Hassiacum In Complex With Sam And Glycerol.
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: SAH, GOL, MG, CL |
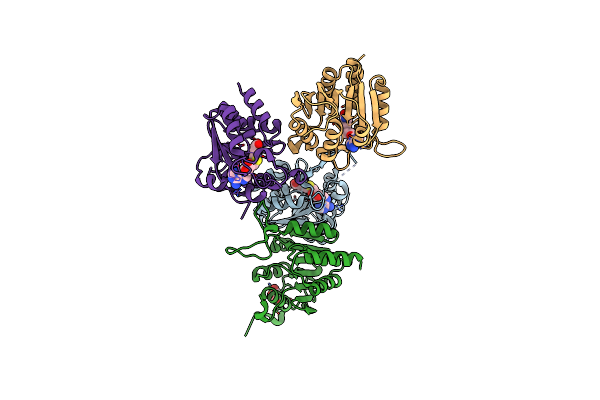 |
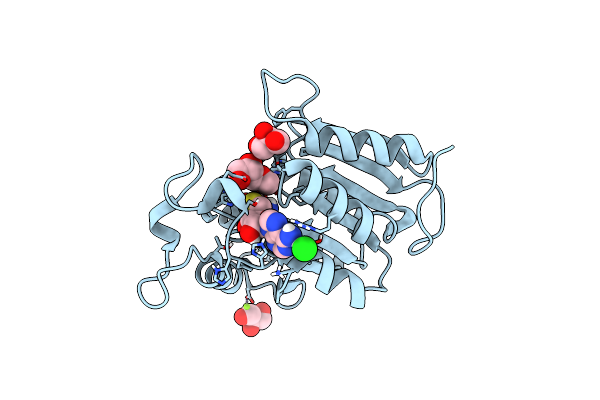
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: SAH, GOL |
 |
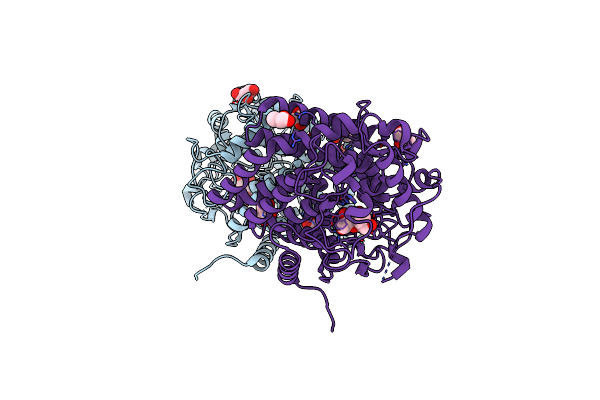
High Resolution Structure Of Met1 From Mycobacterium Hassiacum In Complex With 3-Methoxy-1,2-Propanediol.
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: GOL, FW5, SAH, MG, CL |
 |
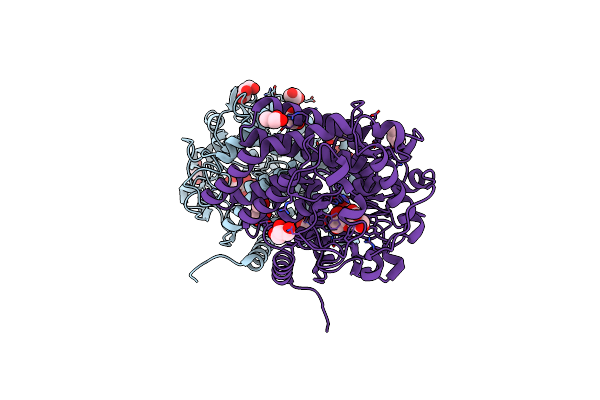
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase(Mhggh) E419A Variant In Complex With Glucosylglycerol
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: GOL, A0K |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) D182A Variant In Complex With Glucosylglycolate
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: GOL, 9YW, SER |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) E419A Variant In Complex With Glucosylglycolate
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: GOL, 9YW, SER |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) - Semet Derivative
Organism: Mycobacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2018-08-08 Classification: HYDROLASE Ligands: GOL, SER |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) In Complex With Serine And Glycerol
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2018-08-08 Classification: HYDROLASE Ligands: GOL, SER |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) D182A Variant In Complex With Serine And Glycerol
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-08-08 Classification: HYDROLASE Ligands: GOL, SER |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) E419A Variant In Complex With Serine And Glycerol
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2018-08-08 Classification: HYDROLASE Ligands: GOL, SER, PEG |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) D43A Variant In Complex With Serine And Glycerol
Organism: Mycobacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2018-08-08 Classification: HYDROLASE Ligands: GOL, SER, GLY |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) D182A Variant In Complex With Glucosylglycerate
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2018-08-08 Classification: HYDROLASE Ligands: GOL, PEG, PGE, 9WN |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) D182A Variant In Complex With Mannosylglycerate
Organism: Mycobacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-08-08 Classification: HYDROLASE Ligands: GOL, PEG, GLY, 2M8 |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) E419A Variant In Complex With Glucosylglycerate
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2018-08-08 Classification: HYDROLASE Ligands: 9WN, GOL, PEG |

