Search Count: 38
 |
The Crystal Structure Of Recombinant Chloroperoxidase Expressed In Aspergillus Niger
Organism: Leptoxyphium fumago
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: MN, HEM, NAG, MAN, IOD |
 |
Crystal Structure Of C-As Lyase With Mutation K105A And Substrate Roxarsone
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9)
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2018-03-21 Classification: OXIDOREDUCTASE Ligands: RXO, FE, CL |
 |
Crystal Structure Of C-As Lyase With Mutations Y100H And V102F (Monoclinic Form)
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / ncimb 10081)
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2017-01-18 Classification: LYASE |
 |
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / ncimb 10081)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2016-09-21 Classification: LYASE |
 |
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / ncimb 10081)
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2016-09-21 Classification: LYASE |
 |
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / ncimb 10081)
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2016-09-07 Classification: LYASE Ligands: FE, NA, CL |
 |
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / ncimb 10081)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2016-07-13 Classification: OXIDOREDUCTASE Ligands: NI |
 |
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / ncimb 10081)
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2016-07-13 Classification: OXIDOREDUCTASE Ligands: NI, NA |
 |
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / ncimb 10081)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-07-13 Classification: LYASE Ligands: CO, GOL, NA |
 |
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / ncimb 10081)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-07-13 Classification: LYASE Ligands: NI, CL, BME |
 |
Crystal Structure Of Escherichia Coli Glutaredoxin 2 Complex With Glutathione
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-05-28 Classification: ELECTRON TRANSPORT Ligands: GSH, ACY |
 |
Crystal Structure Of Escherichia Coli Glutraredoxin 2 C9S/C12S Mutant Without Glutathione
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-05-21 Classification: ELECTRON TRANSPORT Ligands: TRS |
 |
Method: X-RAY DIFFRACTION
Resolution:1.86 Å Release Date: 2013-03-20 Classification: DNA Ligands: BA |
 |
Method: X-RAY DIFFRACTION
Resolution:1.67 Å Release Date: 2013-03-13 Classification: DNA Ligands: BA |
 |
Method: X-RAY DIFFRACTION
Resolution:1.72 Å Release Date: 2013-03-13 Classification: DNA Ligands: BA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2012-11-28 Classification: DNA Ligands: MN |
 |
Method: X-RAY DIFFRACTION
Resolution:1.76 Å Release Date: 2012-11-28 Classification: DNA Ligands: MN |
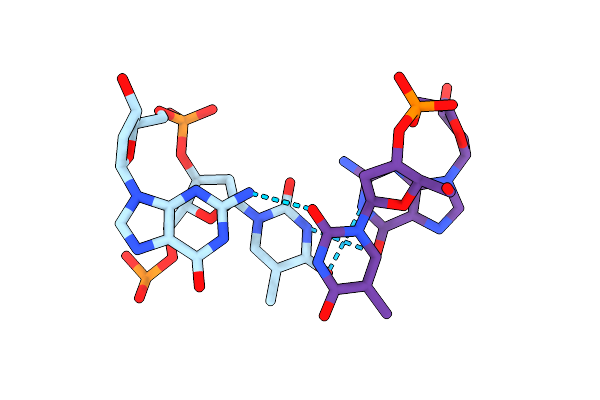 |
X-Ray Diffraction Studies Of Ring Crystals Obtained For D(Cacgcg).D(Cgcgtg): Stage (Ii) Hexagonal Plates With Spots
Method: X-RAY DIFFRACTION
Resolution:3.01 Å Release Date: 2012-07-25 Classification: DNA Ligands: HOH |
 |
X-Ray Diffraction Studies Of Ring Crystals Obtained For D(Cacgcg).D(Cgcgtg): Stage (Iii) Hexagonal Plates With Intense Spots And A Depression
|
 |
X-Ray Diffraction Studies Of Ring Crystals Obtained For D(Cacgcg).D(Cgcgtg): Stage (Iv) Hexagonal Rings
Method: X-RAY DIFFRACTION
Resolution:3.24 Å Release Date: 2012-07-25 Classification: DNA Ligands: HOH |

