Search Count: 48
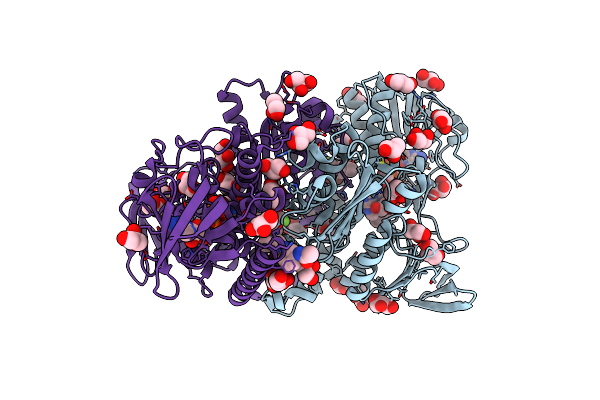 |
Co-Crystal Structure Of Optimized Analog Tdi-13537 Provided New Insights Into The Potency Determinants Of The Sulfonamide Inhibitor Series
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-01-03 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, U4I, GOL |
 |
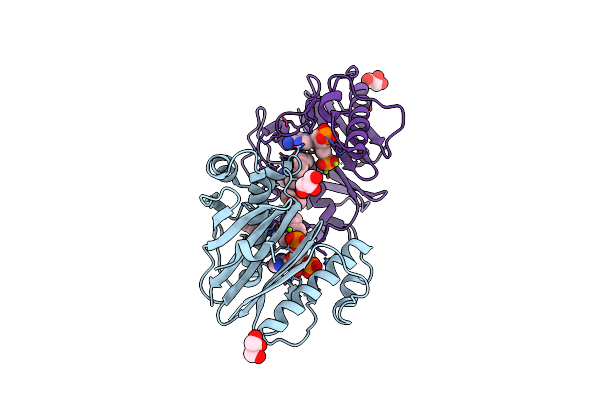
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-02-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: COA, MG, 0UD, DMS |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2022-02-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: COA, 10I, MG, GOL, DMS |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-02-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: COA, 17I, MG, GOL, DMS |
 |
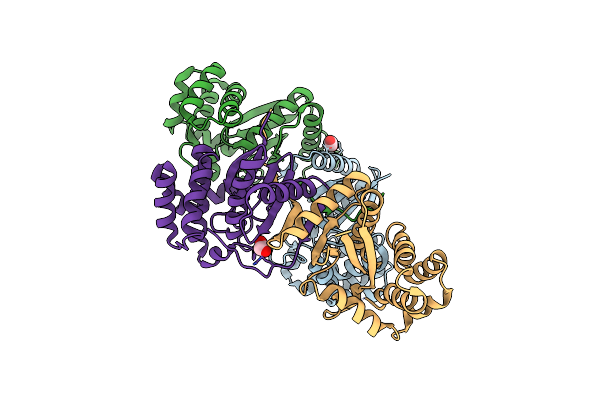
Macrocyclic Peptides Tdi5575 That Selectively Inhibit The Mycobacterium Tuberculosis Proteasome
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2021-04-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: U5Y, CIT, DMF |
 |
Single-Particle Cryo-Em Structure Of Plasmodium Falciparum Chloroquine Resistance Transporter (Pfcrt) 7G8 Isoform
Organism: Plasmodium falciparum (isolate 7g8), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-12-04 Classification: MEMBRANE PROTEIN Ligands: Y01 |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-03-14 Classification: OXIDOREDUCTASE Ligands: EDO, CL |
 |
Organism: Streptomyces sp. bc16019
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-10-12 Classification: HYDROLASE Ligands: CL, 1PE, GOL |
 |
Organism: Streptomyces sp. bc16019
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2016-10-05 Classification: HYDROLASE Ligands: MN, BCT |
 |
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-02-17 Classification: TRANSFERASE Ligands: ZN, DSL, MPG, CL, PO4, PC, EPE |
 |
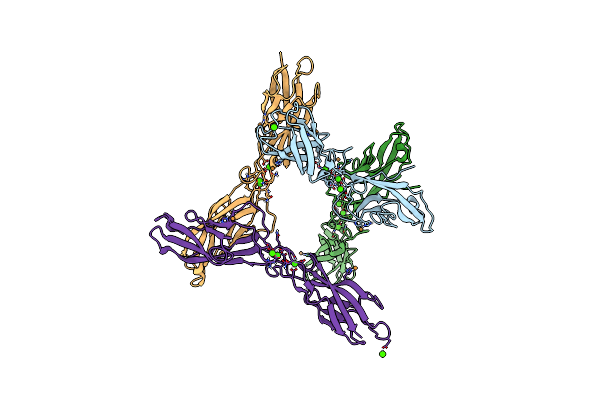
Crystal Structure Of Arnt From Cupriavidus Metallidurans Bound To Undecaprenyl Phosphate
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-02-17 Classification: TRANSFERASE Ligands: MPG, EPE, PO4, CL, 5TR |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2014-09-24 Classification: CELL ADHESION Ligands: CA, CU |
 |
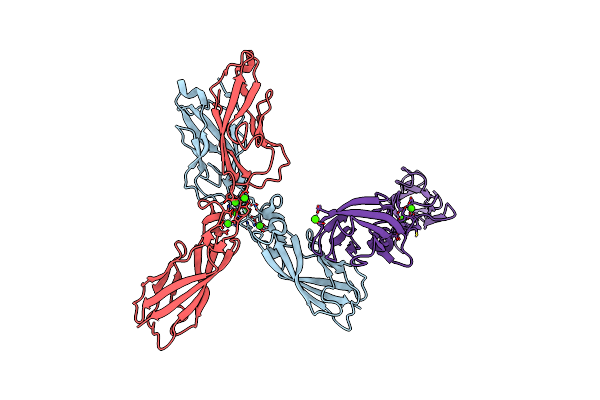
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2014-09-24 Classification: CELL ADHESION Ligands: CA |
 |
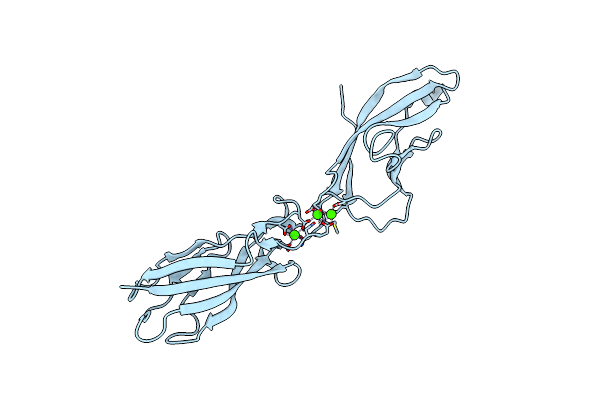
Crystal Structure Of Mouse N-Cadherin Ec1-2 With Aa Insertion Between Residues 2 And 3
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-09-24 Classification: CELL ADHESION Ligands: CA |
 |
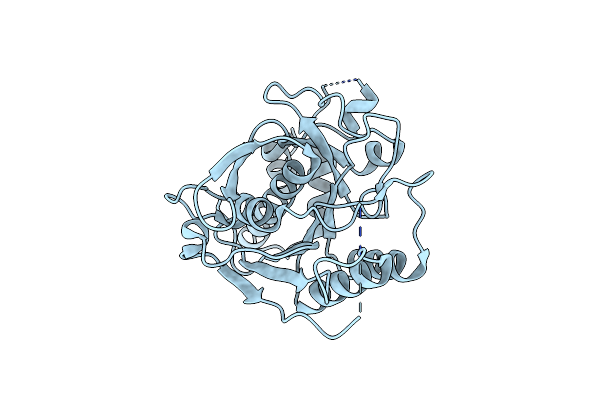
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2014-09-24 Classification: CELL ADHESION Ligands: CA |
 |
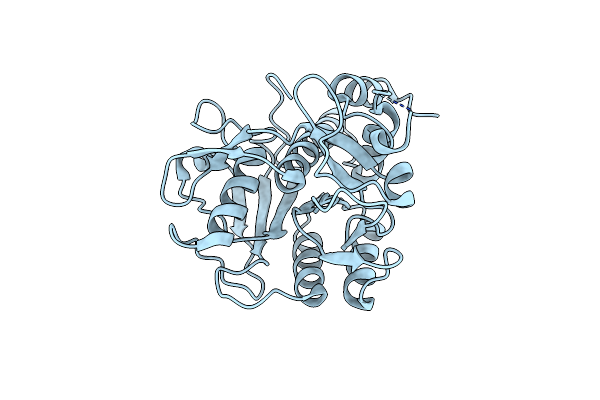
Organism: Prochloron didemni
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-08-29 Classification: HYDROLASE |
 |
Organism: Prochloron didemni
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2012-08-29 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-08-22 Classification: CELL ADHESION Ligands: FMT |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2012-08-22 Classification: CELL ADHESION Ligands: CD |
 |
Crystal Structure Of Mouse Nectin-2 Extracellular Fragment D1-D2, 2Nd Crystal Form
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2012-08-22 Classification: CELL ADHESION Ligands: NAG |

