Search Count: 17
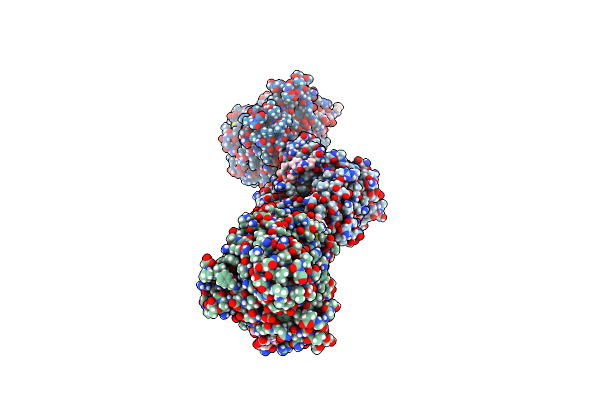 |
Crystal Structure Of Yeast Clathrin Heavy Chain N-Terminal Domain Bound To Epsin-5 Peptide (Lidl)
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-11-13 Classification: ENDOCYTOSIS |
 |
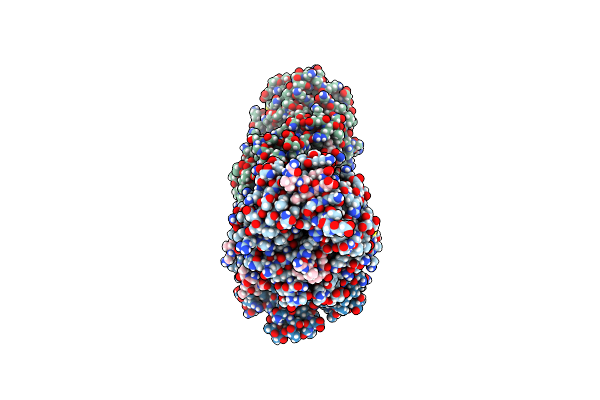
Crystal Structure Of Yeast Clathrin Heavy Chain N-Terminal Domain Bound To Yap1801 Peptide (Lidm)
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-11-13 Classification: ENDOCYTOSIS Ligands: GOL, CL |
 |
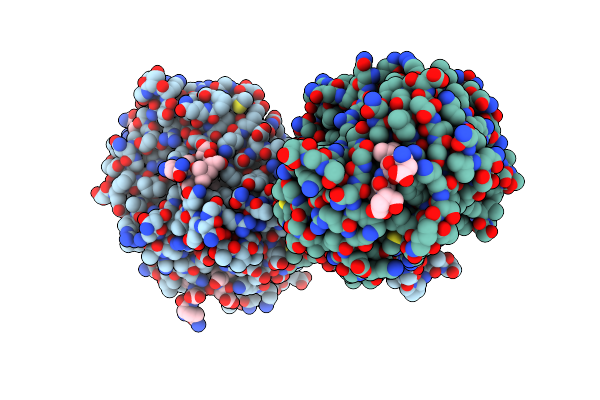
Crystal Structure Of Yeast Clathrin Heavy Chain N-Terminal Domain Bound To Epsin-2 Peptide (Lidl)
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-11-13 Classification: ENDOCYTOSIS Ligands: GOL, PO4 |
 |
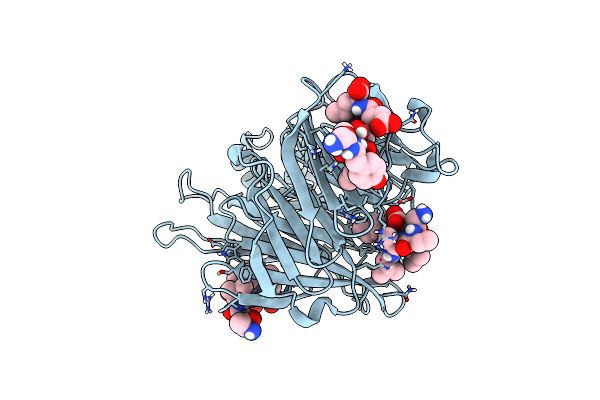
Crystal Structure Of Yeast Clathrin Heavy Chain N-Terminal Domain Bound To Apl2/Ap-1 Beta Peptide (Lldl)
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-11-13 Classification: ENDOCYTOSIS |
 |
Crystal Structure Of Yeast Clathrin Heavy Chain N-Terminal Domain Bound To Epsin-1 Peptide (Lidl)
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-11-13 Classification: ENDOCYTOSIS |
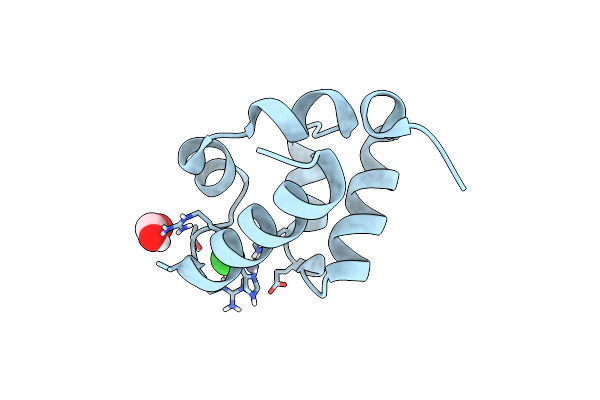 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2022-11-30 Classification: METAL BINDING PROTEIN Ligands: FMT, CL, ZN |
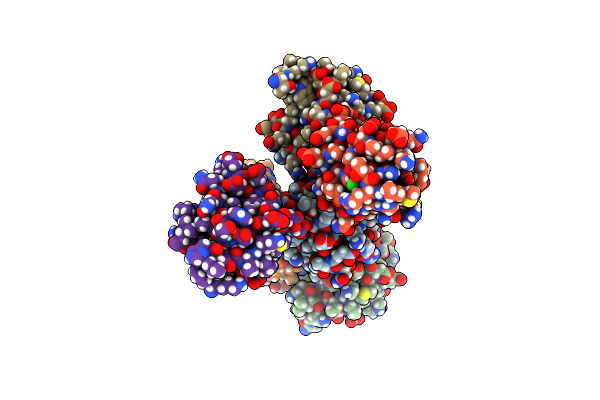 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-11-30 Classification: METAL BINDING PROTEIN Ligands: BTB, PEG, CL |
 |
Organism: Uperoleia mjobergii
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: PROTEIN FIBRIL |
 |
Organism: Ranoidea raniformis
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: PROTEIN FIBRIL |
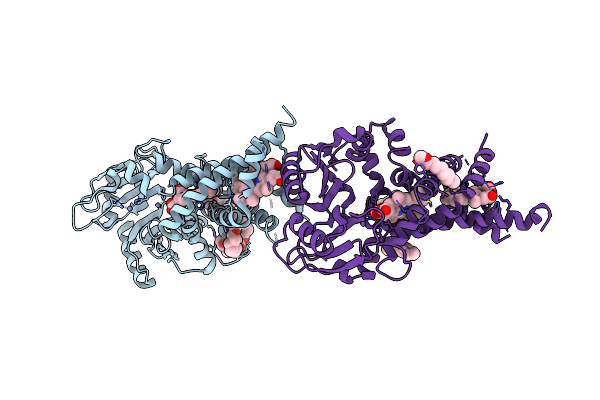 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2021-09-01 Classification: MEMBRANE PROTEIN Ligands: HEM, PE5, DMU, ZN |
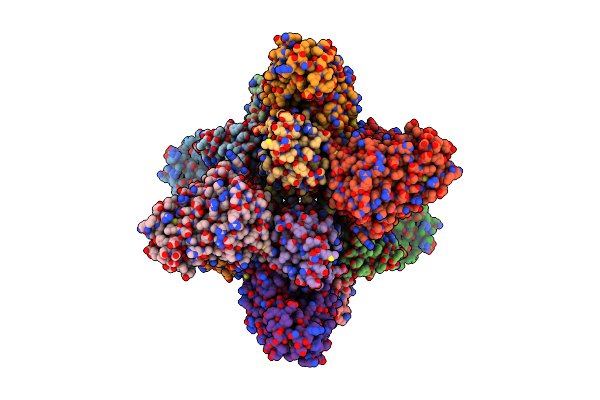 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2021-05-05 Classification: CELL ADHESION Ligands: PIO |
 |
Structure Of The Ferrioxamine B Transporter Foxa From Pseudomonas Aeruginosa In Complex With Ferrioxamine B
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: 0UE, BNG, GOL, SO4, NA |
 |
Structure Of The Ferrioxamine B Transporter Foxa From Pseudomonas Aeruginosa In Complex With Ferrioxamine B And A C-Terminal Tonb Fragment
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: 0UE |
 |
Structure Of The Ferrioxamine B Transporter Foxa From Pseudomonas Aeruginosa, Apo State
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: NI, GOL, C8E, FTT, SO4, NA |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2016-07-06 Classification: RNA BINDING PROTEIN |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-07-06 Classification: RNA BINDING PROTEIN |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-08-29 Classification: TRANSLATION |

