Search Count: 25
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: DE NOVO PROTEIN Ligands: CD, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: DE NOVO PROTEIN |
 |
High-Resolution Structure Of The Siderophore Periplasmic Binding Protein Ftsb From Streptococcus Pyogenes
Organism: Streptococcus pyogenes ssi-1
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2024-10-09 Classification: METAL BINDING PROTEIN Ligands: P33, PEG, EDO, P6G, ZN, NA, CL |
 |
Structure Of The Siderophore Periplasmic Binding Protein Ftsb From Streptococcus Pyogenes With Ferrichrome Bound
Organism: Streptococcus pyogenes ssi-1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-10-09 Classification: METAL BINDING PROTEIN Ligands: FCE |
 |
Structure Of The Siderophore Periplasmic Binding Protein Ftsb From Streptococcus Pyogenes With Bisucaberin Bound
Organism: Streptococcus pyogenes ssi-1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-09 Classification: METAL BINDING PROTEIN Ligands: OX8, FE |
 |
High-Resolution Structure Of The Siderophore Periplasmic Binding Protein Ftsb From Streptococcus Pyogenes With Ferrioxamine E Bound
Organism: Streptococcus pyogenes ssi-1
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2024-10-09 Classification: METAL BINDING PROTEIN Ligands: 6L0, FE, ZN, NA, GOL, EDO |
 |
High-Resolution Structure Of The Siderophore Periplasmic Binding Protein Ftsb From Streptococcus Pyogenes With Ferrioxamine B
Organism: Streptococcus pyogenes ssi-1
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-10-09 Classification: METAL BINDING PROTEIN Ligands: 0UE, ZN, EDO, 03S, NA |
 |
High-Resolution Structure Of The Siderophore Periplasmic Binding Protein Ftsb Mutant Y137A From Streptococcus Pyogenes
Organism: Streptococcus pyogenes ssi-1
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-10-09 Classification: METAL BINDING PROTEIN Ligands: P33, PEG, ZN, SO4, CL, NA |
 |
Structure Of The Siderophore Periplasmic Binding Protein Ftsb Mutant Y137A From Streptococcus Pyogenes With Ferrioxamine E Bound
Organism: Streptococcus pyogenes ssi-1
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-10-09 Classification: METAL BINDING PROTEIN Ligands: 6L0, FE, GOL |
 |
High-Resolution Structure Of The Siderophore Periplasmic Binding Protein Ftsb From Streptococcus Pyogenes With Ferrioxamine E Bound (Crystal Form 2)
Organism: Streptococcus pyogenes ssi-1
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-10-09 Classification: METAL BINDING PROTEIN Ligands: 6L0, FE, CL |
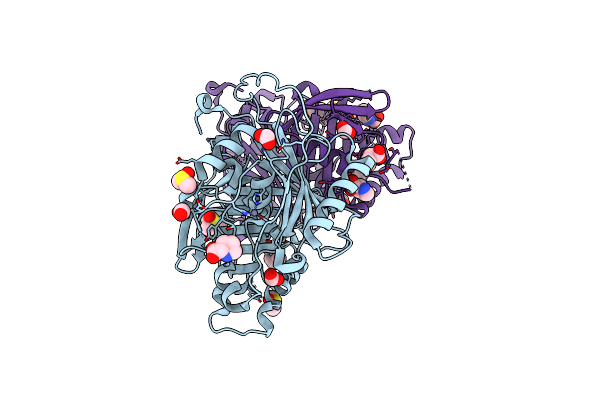 |
Crystal Structure Of Profragilysin-3 (Probft-3) From Bacteroides Fragilis At 1.85 A Resolution.
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ZN, MG, DMS, PRO, FMT |
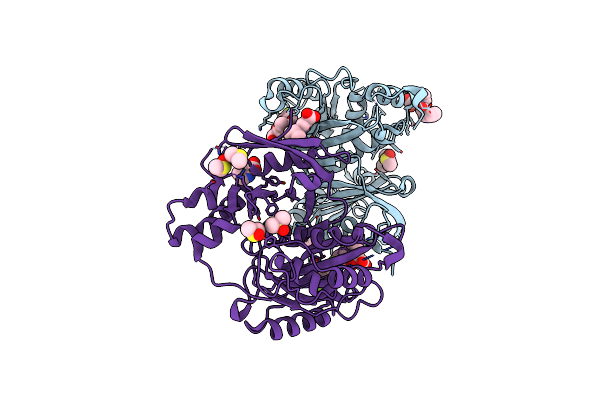 |
Crystal Structure Of Profragilysin-3 (Probft-3) From Bacteroides Fragilis In Complex With Flumequine
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ZN, 7X9, CL, PRO, DMS |
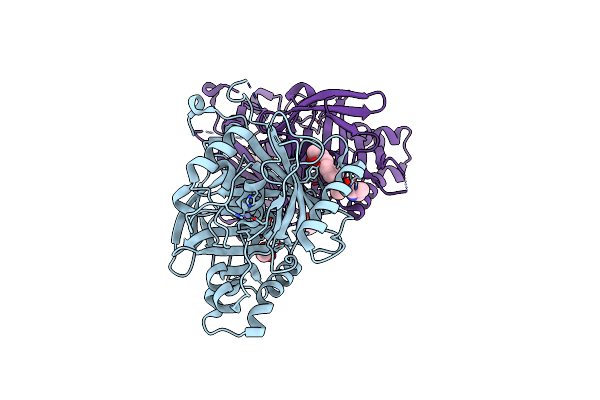 |
Crystal Structure Of Profragilysin-3 (Probft-3) From Bacteroides Fragilis In Complex With Foliosidine In P212121.
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ZN, 7WK, ACT, PGE, PRO |
 |
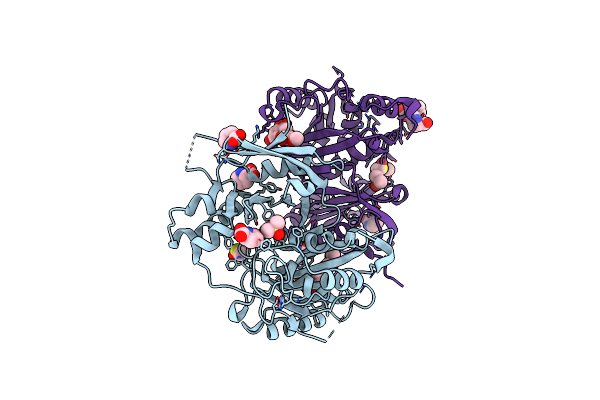
Crystal Structure Of Profragilysin-3 (Probft-3) From Bacteroides Fragilis In Complex With Foliosidine In P41212.
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ZN, 7WK, CL, DMS, PRO, PEG, EDO, PGE |
 |
Crystal Structure Of Profragilysin-3 (Probft-3) From Bacteroides Fragilis In Complex With Hesperetin.
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ZN, 6JP, DMS, PRO |
 |
Organism: Nostoc sp. (strain atcc 29151 / pcc 7119)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-08-02 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Organism: Bothrops brazili
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2013-11-20 Classification: TOXIN |

