Search Count: 13
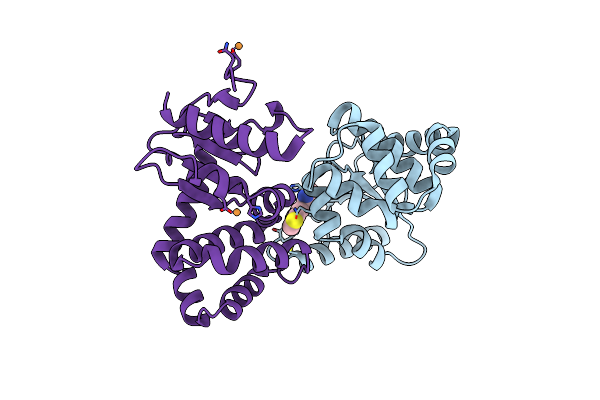 |
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001897 (Compound 1)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 5VA, CU |
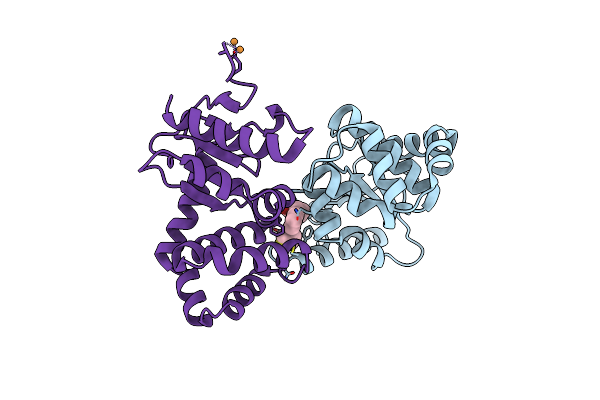 |
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001877 (Compound 39)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 648, CU |
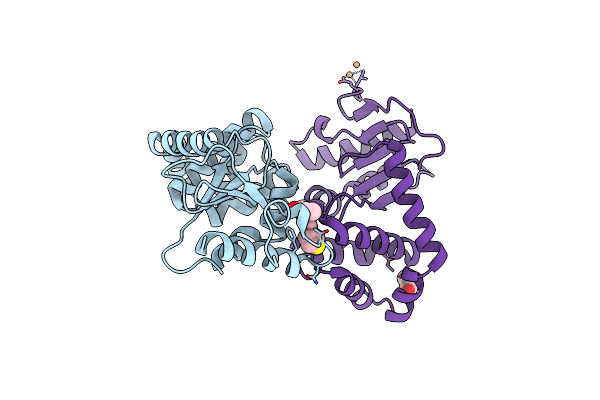 |
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001886 (Compound 38)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 5VB, CU, GOL |
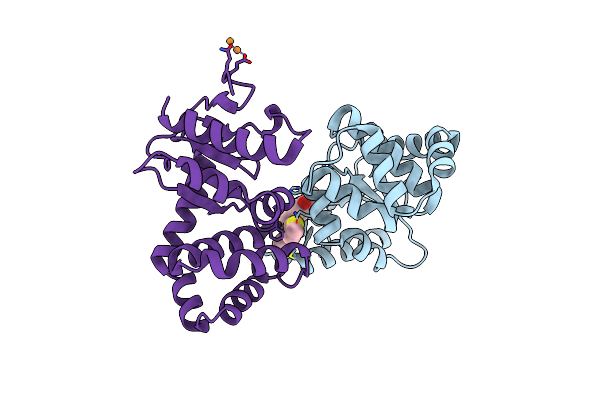 |
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001896 (Compound 72)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 62J, CU |
 |
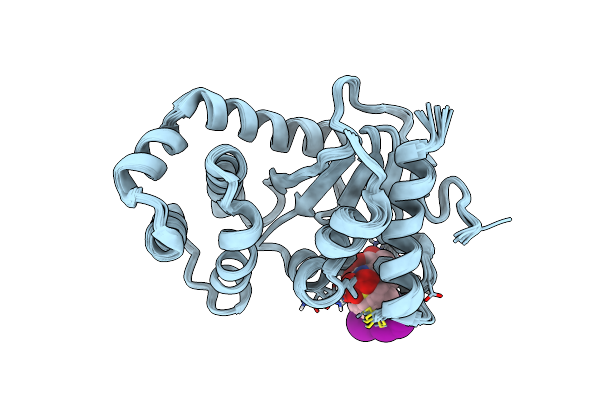
Model Structure Of The Oxidized Padsba1 And 3-[(2-Methylbenzyl)Sulfanyl]-4H-1,2,4-Triazol-4-Amine Complex
Organism: Pseudomonas aeruginosa
Method: SOLUTION NMR Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: 1YO |
 |
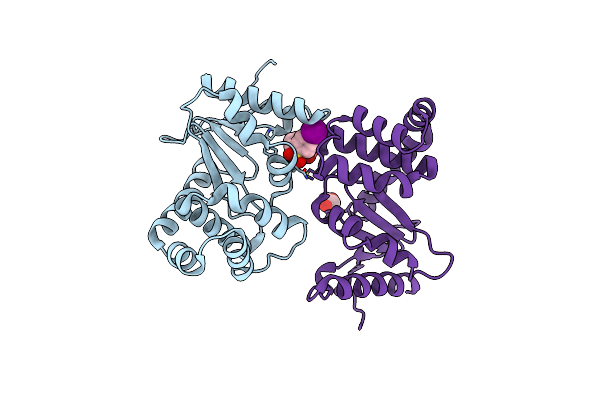
Organism: Escherichia coli k-12
Method: SOLUTION NMR Release Date: 2017-02-08 Classification: OXIDOREDUCTASES Ligands: SFQ |
 |
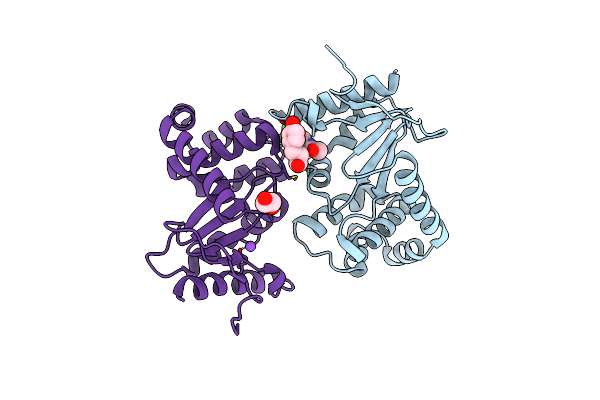
Crystal Structure Of Pseudomonas Aeruginosa Dsba E82I In Complex With Mips-0000851 (3-[(2-Methylbenzyl)Sulfanyl]-4H-1,2,4-Triazol-4-Amine)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-10-05 Classification: OXIDOREDUCTASE Ligands: 1YO, GOL, MES |
 |
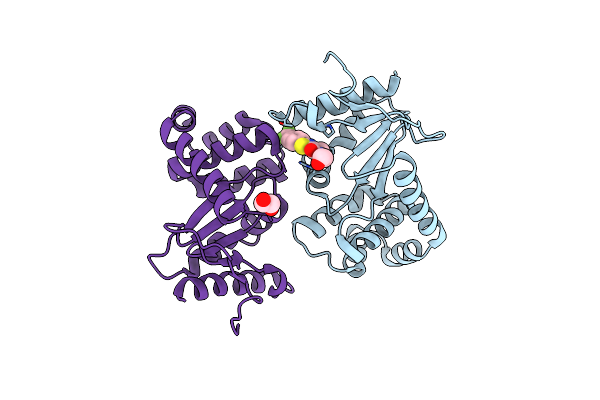
Crystal Structure Of E.Coli Dsba In Complex With 2-(4-Iodophenylsulfonamido) Benzoic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2016-05-11 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: SFQ, EDO |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2015-01-21 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: WEF, EDO, NA |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-01-21 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: EDO, EG6 |
 |
Crystal Structure Of E.Coli Dsba Co-Crystallised In Complex With Compound 4
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-01-21 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: EDO, WF4 |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2015-01-21 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: WF4, CU, GOL, EDO |
 |
Organism: Pseudomonas aeruginosa
Method: SOLUTION NMR Release Date: 2014-11-12 Classification: OXIDOREDUCTASE |

