Search Count: 148
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: SUGAR BINDING PROTEIN Ligands: CA, A1IZM, 1PE, EDO, CL, PG4, 9YU, ACT, PEG |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: SUGAR BINDING PROTEIN Ligands: CA, A1I1G |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-04-16 Classification: SUGAR BINDING PROTEIN Ligands: CA, SO4, R7E |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-04-16 Classification: SUGAR BINDING PROTEIN Ligands: CA, R7E, A1IH4 |
 |
N Terminal Domain Of Bc2L-C Lectin In Complex With N-(Beta-L-Fucopyranosyl)-Biphenyl-3-Carboxamide
Organism: Burkholderia cenocepacia j2315
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-04-16 Classification: SUGAR BINDING PROTEIN Ligands: MJO, PG4, 1PE |
 |
Crystal Structure Of Trichuris Suis Beta-N-Acetyl-D-Hexosaminidase - Hex-2 In Apo Form
Organism: Trichuris suis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2024-06-26 Classification: HYDROLASE Ligands: ZN |
 |
Structure Of The N-Terminal Domain Of Cma In Complex With N-Acetyllactosamine
Organism: Cucumis melo
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2023-12-27 Classification: SUGAR BINDING PROTEIN Ligands: CD |
 |
Structure Of The N-Terminal Domain Of Cma From Cucumis Melo In Complex With N-Acetylgalactosamine
Organism: Cucumis melo
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-12-27 Classification: SUGAR BINDING PROTEIN Ligands: CD, NGA, A2G |
 |
Leca From Pseudomonas Aeruginosa In Complex With Tolcapone (Cas: 134308-13-7)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2023-07-19 Classification: SUGAR BINDING PROTEIN Ligands: CA, TCW |
 |
Structure Of The N-Terminal Domain Of Bc2L-C Lectin (1-131) In Complex With A Synthetic Beta-Fucosylamide
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-02-08 Classification: SUGAR BINDING PROTEIN Ligands: R7E |
 |
Structure Of The Leca Lectin From Pseudomonas Aeruginosa In Complex With A Biaryl-Thiogalactoside
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-01-18 Classification: SUGAR BINDING PROTEIN Ligands: CA, IEC, EDO |
 |
Structure Of The Leca Lectin From Pseudomonas Aeruginosa In Complex With A Biaryl-Thiogalactoside
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-01-18 Classification: SUGAR BINDING PROTEIN Ligands: CA, IE3, EDO |
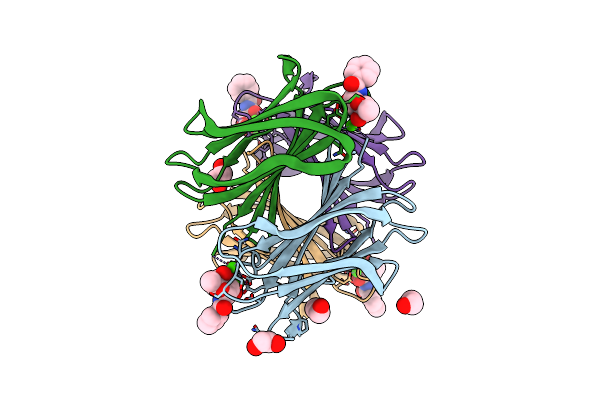 |
Structure Of The Lecb Lectin From Pseudomonas Aeruginosa Strain Pao1 In Complex With N-(Alpha-L-Fucopyranosyl)Benzamide (6)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-11-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, M9I, EDO |
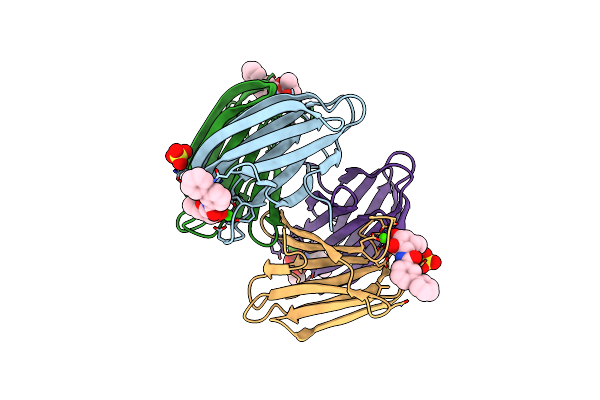 |
Structure Of The Lecb Lectin From Pseudomonas Aeruginosa Strain Pao1 In Complex With N-(Beta-L-Fucopyranosyl)-Biphenyl-3-Carboxamide (4I)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-11-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, MJO, SO4 |
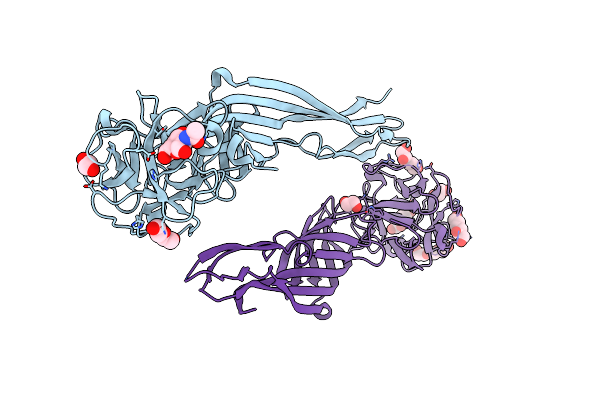 |
Se-M Variant Of B-Trefoil Lectin From Salpingoeca Rosetta In Complex With Galnac
Organism: Salpingoeca rosetta
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN Ligands: NGA, EDO, PEG, 1JW, PGE |
 |
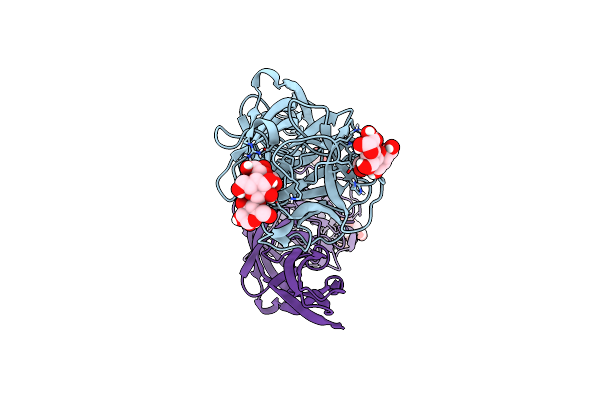
Organism: Salpingoeca rosetta
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN Ligands: A2G, NGA, CL, NA |
 |
Organism: Salpingoeca rosetta
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN Ligands: EDO |
 |
Structure Of The N-Terminal Domain Of Bc2L-C Lectin (1-131) In Complex With Lewis Y Antigen
Organism: Burkholderia cenocepacia (strain atcc baa-245 / dsm 16553 / lmg 16656 / nctc 13227 / j2315 / cf5610)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-06-01 Classification: SUGAR BINDING PROTEIN |
 |
Structure Of The N-Terminal Domain Of Bc2L-C Lectin (1-131) In Complex With A Synthetic Beta-C-Fucoside Ligand
Organism: Burkholderia cenocepacia (strain atcc baa-245 / dsm 16553 / lmg 16656 / nctc 13227 / j2315 / cf5610)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-06-01 Classification: SUGAR BINDING PROTEIN Ligands: VJT |
 |
Structure Of The N-Terminal Domain Of Bc2L-C Lectin (1-131) In Complex With A Synthetic Beta-N-Fucoside Ligand
Organism: Burkholderia cenocepacia (strain atcc baa-245 / dsm 16553 / lmg 16656 / nctc 13227 / j2315 / cf5610)
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2022-06-01 Classification: SUGAR BINDING PROTEIN Ligands: VJW |

