Search Count: 57
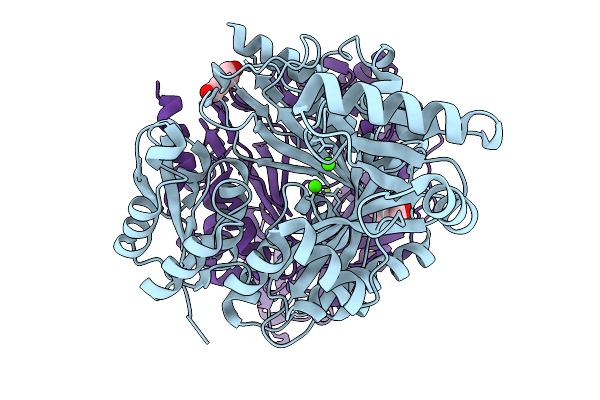 |
Organism: Ankistrodesmus falcatus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LIGASE Ligands: PEG, CA, MG |
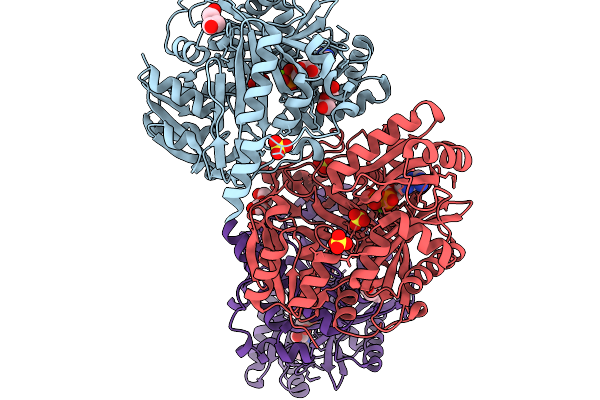 |
Crystal Structure Of Biotin Carboxylase From Ankistrodesmus In Complex With Adp
Organism: Ankistrodesmus falcatus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LIGASE Ligands: MG, ADP, PEG, SO4 |
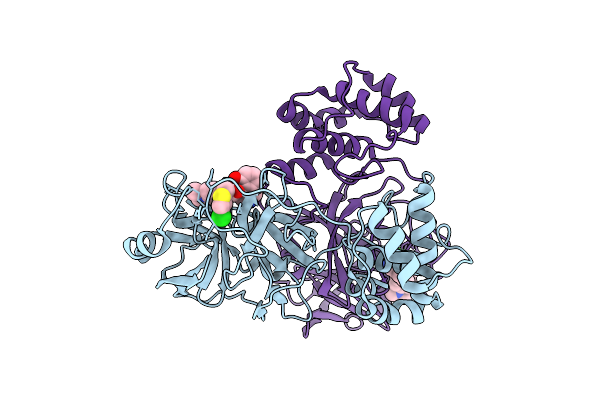 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN, HYDROLASE Ligands: WZK |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN, HYDROLASE |
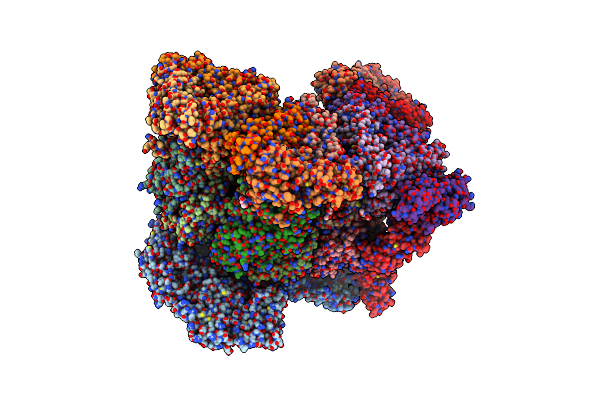 |
Organism: Rotavirus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: VIRUS Ligands: CA, NAG, ZN |
 |
Organism: Rotavirus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: VIRUS Ligands: CA, NAG, ZN |
 |
Cryo-Em Reconstruction Of Vp4 Assembly From Sa11 Rotavirus Non-Tripsinized Triple Layered Particle
|
 |
Cryo-Em Reconstruction Of Vp5*/Vp8* Assembly From Sa11 Rotavirus Tripsinized Triple Layered Particle
|
 |
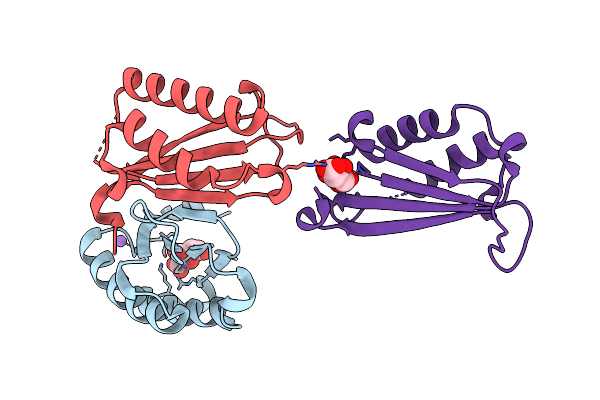
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2024-05-01 Classification: ISOMERASE |
 |
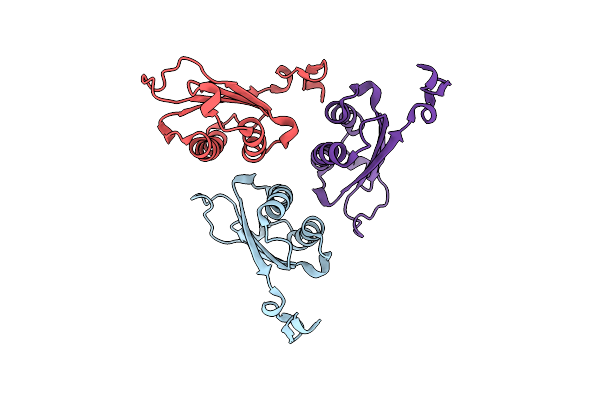
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2024-05-01 Classification: ISOMERASE Ligands: FLC, NA |
 |
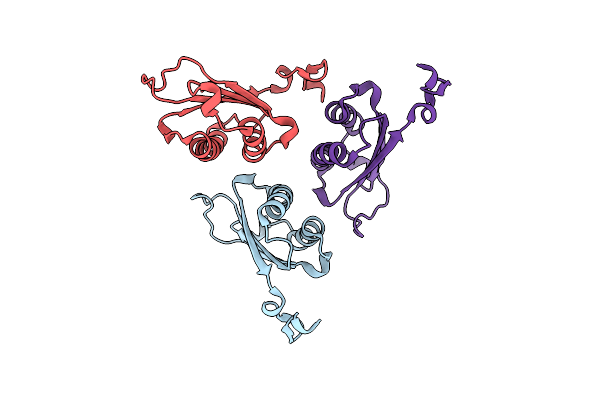
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2024-05-01 Classification: ISOMERASE |
 |
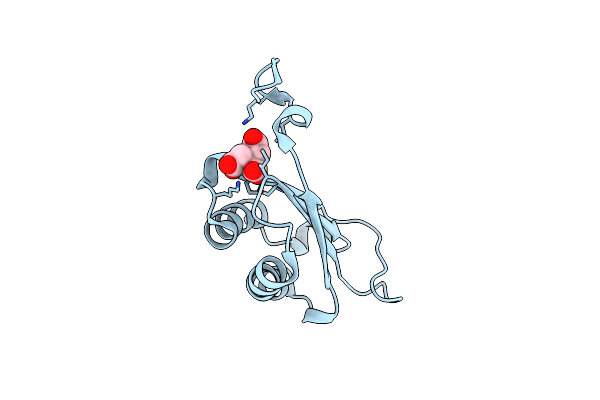
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2024-05-01 Classification: ISOMERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-05-01 Classification: ISOMERASE Ligands: CIT |
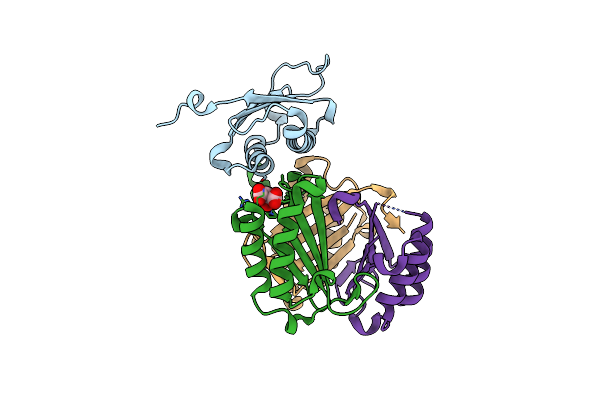 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2024-05-01 Classification: ISOMERASE Ligands: CIT |
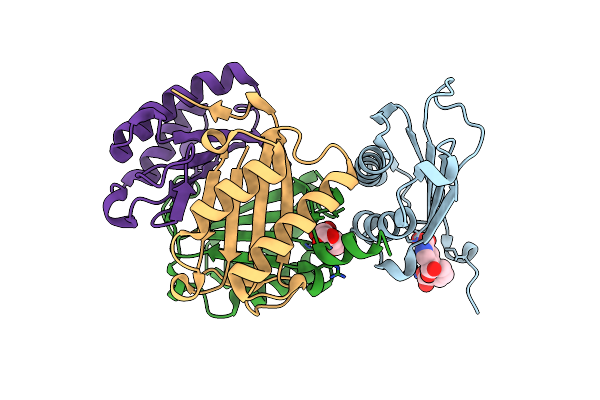 |
Crystal Structure Of V113N Variant Of D-Dopachrome Tautomerase (D-Dt) Bound With 4Cppc
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-05-01 Classification: ISOMERASE Ligands: 7L9, CIT |
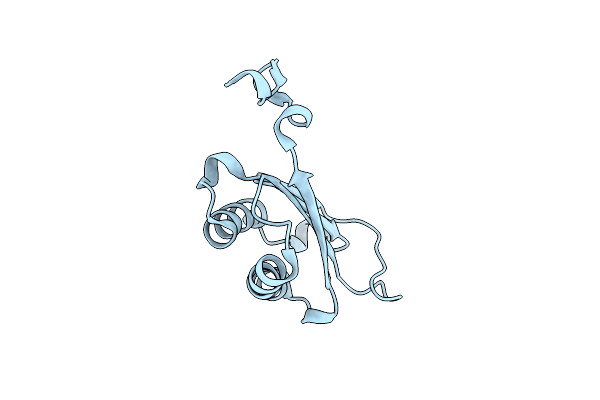 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2024-05-01 Classification: ISOMERASE |
 |
Crystal Structure Of T115A Variant Of D-Dopachrome Tautomerase (D-Dt) Bound To 4Cppc
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2024-05-01 Classification: ISOMERASE Ligands: 7L9 |
 |
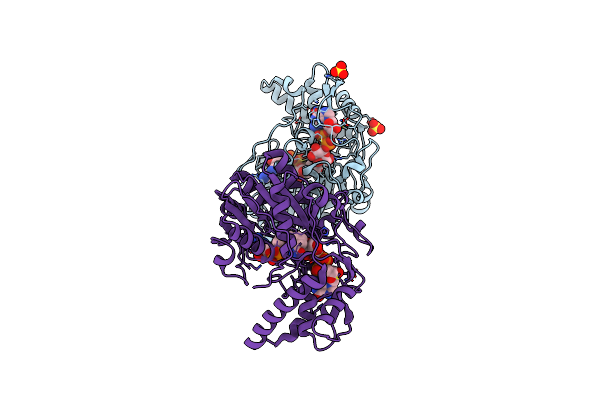
Organism: Myrciaria dubia
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-03-13 Classification: PLANT PROTEIN |
 |
Crystal Structure Of Gdp-Manose 3,5 Epimerase De Myrciaria Dubia In Complex With Substrate, Product And Nad
Organism: Myrciaria dubia
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-03-13 Classification: PLANT PROTEIN Ligands: NAD, SO4, GDD, GDC |
 |
Crystal Structure Of Gdp-Mannose 3,5 Epimerase De Myrciaria Dubia In Complex With Nad
Organism: Myrciaria dubia
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-03-13 Classification: PLANT PROTEIN Ligands: NAD |

