Search Count: 36
 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: PROTEIN BINDING Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: PROTEIN BINDING |
 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: PROTEIN BINDING |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-06-08 Classification: TOXIN Ligands: CL |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-06-08 Classification: TOXIN Ligands: CL |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-06-08 Classification: TOXIN Ligands: CL |
 |
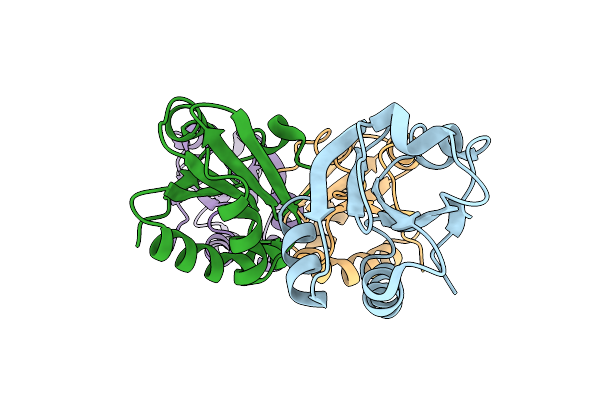
3D Model Of The 3-Rbd Up Single Trimeric Spike Protein Of Sars-Cov2 In The Presence Of Synthetic Peptide Sih-5.
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-04-27 Classification: VIRAL PROTEIN Ligands: NAG |
 |
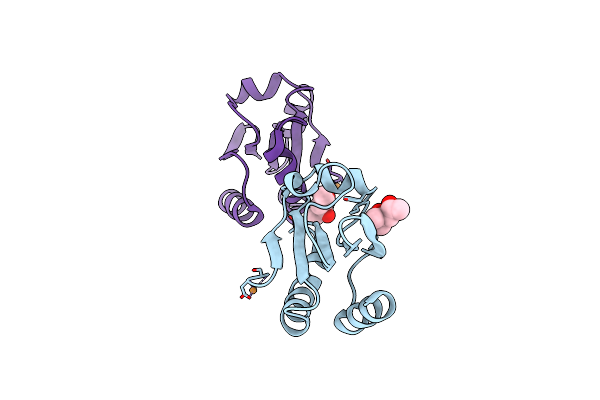
Design Of Disulfide-Linked Thioredoxin Dimers And Multimers Through Analysis Of Crystal Contacts
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-09-11 Classification: ELECTRON TRANSPORT |
 |
Design Of Disulfide-Linked Thioredoxin Dimers And Multimers Through Analysis Of Crystal Contacts
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-09-11 Classification: ELECTRON TRANSPORT Ligands: CU, MPD |
 |
Design Of Disulfide-Linked Thioredoxin Dimers And Multimers Through Analysis Of Crystal Contacts
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-09-11 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-03-29 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-03-29 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-03-29 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-11-13 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2001-06-06 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2001-06-06 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2001-06-06 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2001-06-06 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2000-08-09 Classification: HYDROLASE Ligands: SO4 |
 |
The Role Of Phenylalanine 8 In The Stabilization Of The S Protein-S Peptide Interaction: Packing And Cavities
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 1999-10-20 Classification: HYDROLASE Ligands: SO4 |

