Search Count: 35
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-07-10 Classification: UNKNOWN FUNCTION Ligands: PEG, EDO, NI |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION Ligands: EDO, GOL |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION Ligands: EDO, NA |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION Ligands: EDO, K |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION Ligands: NI, EDO, PEG |
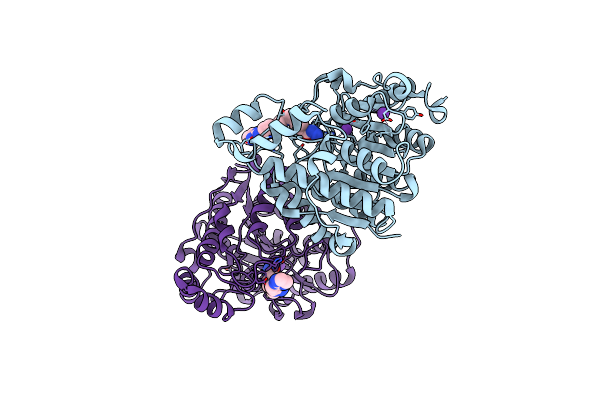 |
Crystal Structure Of Danio Rerio Hdac6 Cd2 In Complex With In Situ Enzymatically Hydrolyzed Dfmo-Based Itf5924
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: LDL, ZN, K |
 |
Organism: Apis mellifera
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2022-07-13 Classification: UNKNOWN FUNCTION Ligands: EDO, MG, NA |
 |
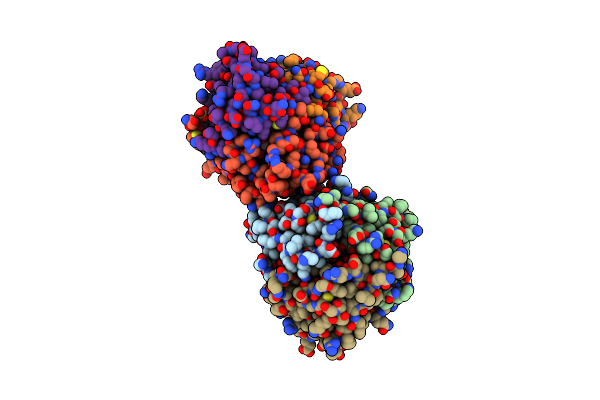
Organism: Salmo salar
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2020-07-29 Classification: UNKNOWN FUNCTION Ligands: ACT, EDO, PEG |
 |
Organism: Salmo salar
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2020-07-29 Classification: UNKNOWN FUNCTION Ligands: ACT, SO4 |
 |
Crystal Structure Of The Pathological N184K Variant Of Calcium-Free Human Gelsolin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2019-11-27 Classification: STRUCTURAL PROTEIN Ligands: CL, GOL, SO4, TRS |
 |
Crystal Structure Of The Pathological G167R Variant Of Calcium-Free Human Gelsolin,
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2019-11-27 Classification: STRUCTURAL PROTEIN Ligands: SO4, GOL |
 |
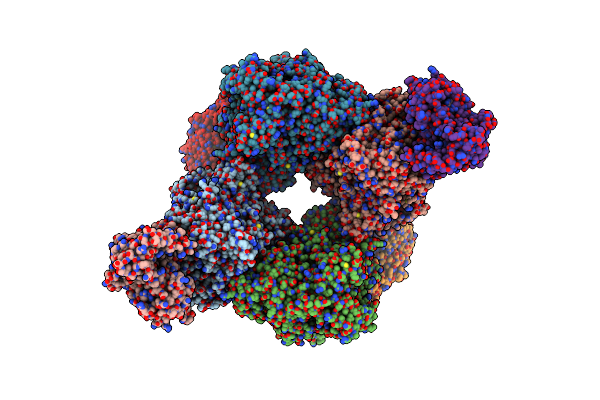
Crystal Structure Of The Pathological D187N Variant Of Calcium-Free Human Gelsolin.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2019-11-27 Classification: STRUCTURAL PROTEIN Ligands: SO4, GOL, NA |
 |
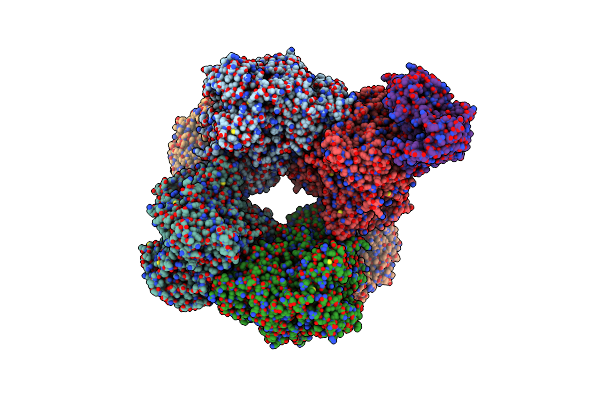
Structure Of Azospirillum Brasilense Glutamate Synthase In A4B4 Oligomeric State.
Organism: Azospirillum brasilense
Method: ELECTRON MICROSCOPY Release Date: 2019-09-11 Classification: FLAVOPROTEIN Ligands: FMN, F3S, SF4, FAD |
 |
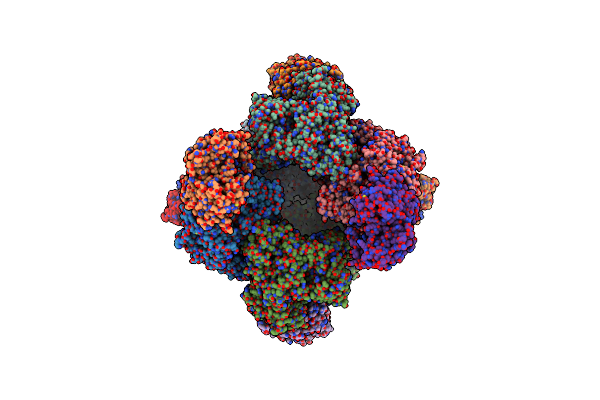
Structure Of Azospirillum Brasilense Glutamate Synthase In A4B3 Oligomeric State
Organism: Azospirillum brasilense
Method: ELECTRON MICROSCOPY Release Date: 2019-09-11 Classification: FLAVOPROTEIN Ligands: FMN, F3S, SF4, FAD |
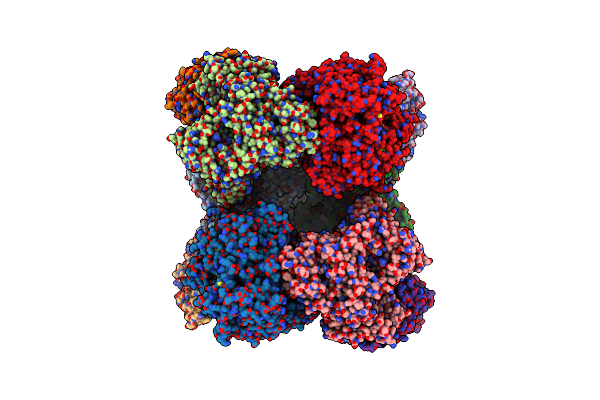 |
Structure Of Azospirillum Brasilense Glutamate Synthase In A6B4 Oligomeric State.
Organism: Azospirillum brasilense
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2019-09-11 Classification: FLAVOPROTEIN Ligands: FMN, F3S, SF4, FAD |
 |
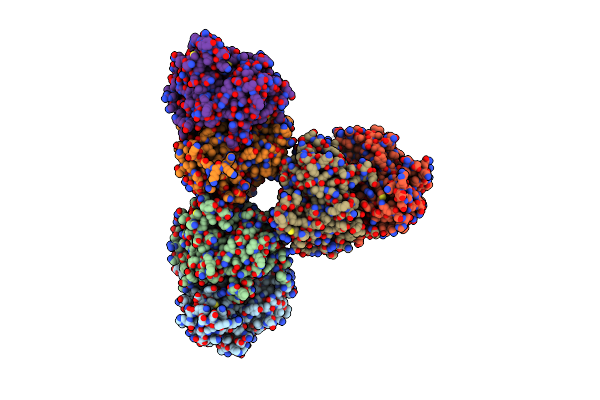
Structure Of Azospirillum Brasilense Glutamate Synthase In A6B6 Oligomeric State.
Organism: Azospirillum brasilense
Method: ELECTRON MICROSCOPY Release Date: 2019-09-11 Classification: FLAVOPROTEIN Ligands: FMN, F3S, SF4, FAD |
 |
Crystal Structure Of The H286L Mutant Of Ferredoxin-Nadp+ Reductase From Plasmodium Falciparum With 2'P-Amp
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2010-01-12 Classification: OXIDOREDUCTASE Ligands: FAD, A2P |

