Search Count: 6
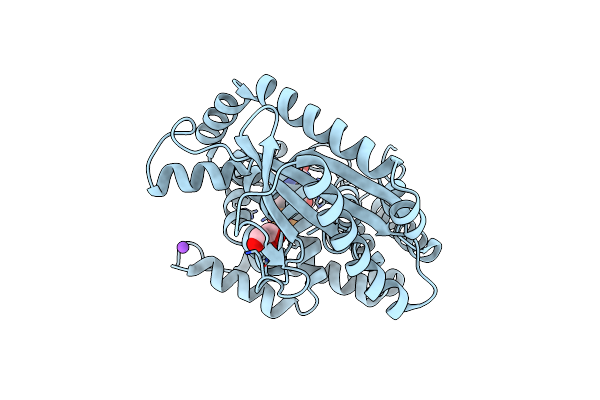 |
Structure Of The N-Terminal Catalytic Region Of T. Thermophilus Rel Bound To Ppgpp
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: G4P, GOL, MN, MG, CL, NA |
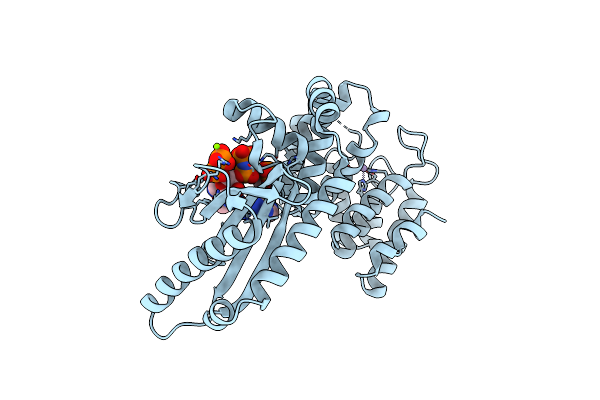 |
Structure Of The Catalytic Domain Of T. Thermophilus Rel In Complex With Amp And Ppgpp
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: MN, AMP, GN3, TMO, MG |
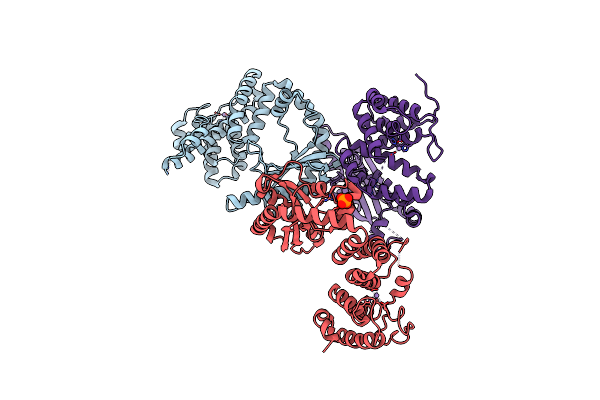 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: MN, CL, PO4, NA |
 |
Structure Of Phosphorylated Translation Elongation Factor Ef-Tu From E. Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-12-20 Classification: HYDROLASE Ligands: MG, GDP |
 |
Organism: Escherichia coli hs
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2017-12-20 Classification: HYDROLASE Ligands: MG, GDP, ACT, EPE, BME, CL |
 |
Organism: Escherichia coli o9:h4 (strain hs)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-12-20 Classification: HYDROLASE Ligands: MG, GDP |

