Search Count: 11
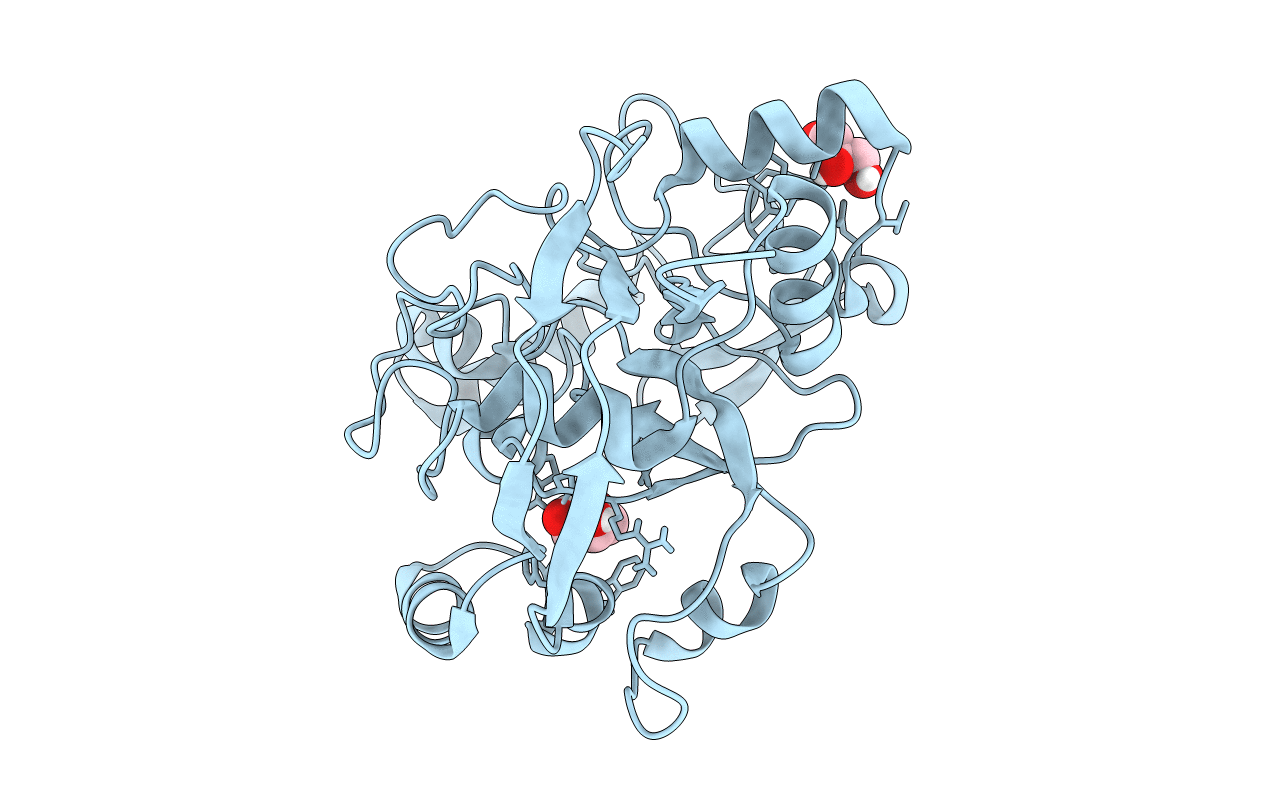 |
Structure Of Calu17 From The Calicheamicin Biosynthesis Pathway Of Micromonospora Echinospora
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-07-28 Classification: UNKNOWN FUNCTION Ligands: GOL |
 |
Structure Of Calu17 From The Calicheamicin Biosynthesis Pathway Of Micromonospora Echinospora
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2021-07-28 Classification: UNKNOWN FUNCTION Ligands: CA, GOL, MG, CL |
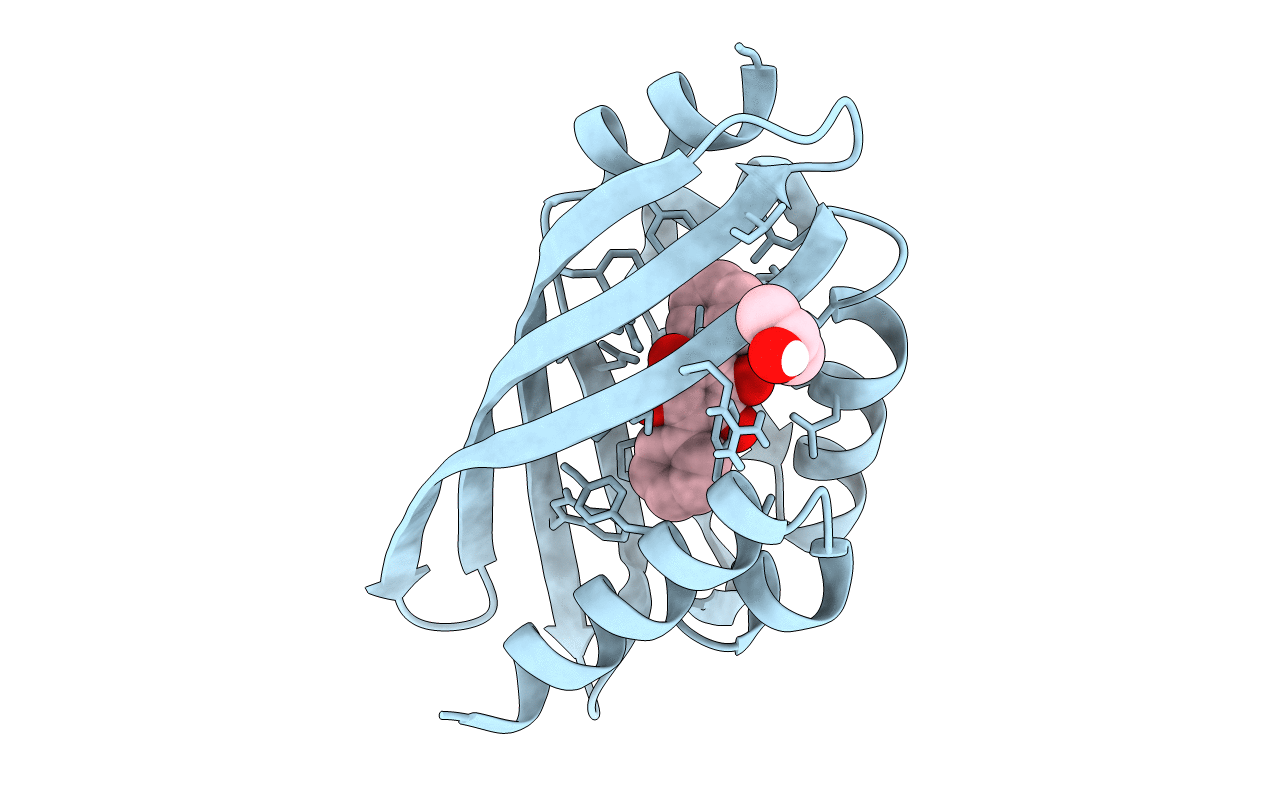 |
Organism: Streptomyces sp. rm-5-8
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-04-07 Classification: BIOSYNTHETIC PROTEIN Ligands: U07, IPA |
 |
Structure Of Calu17 From The Calicheamicin Biosynthesis Pathway Of Micromonospora Echinospora
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-17 Classification: UNKNOWN FUNCTION Ligands: MG, GOL |
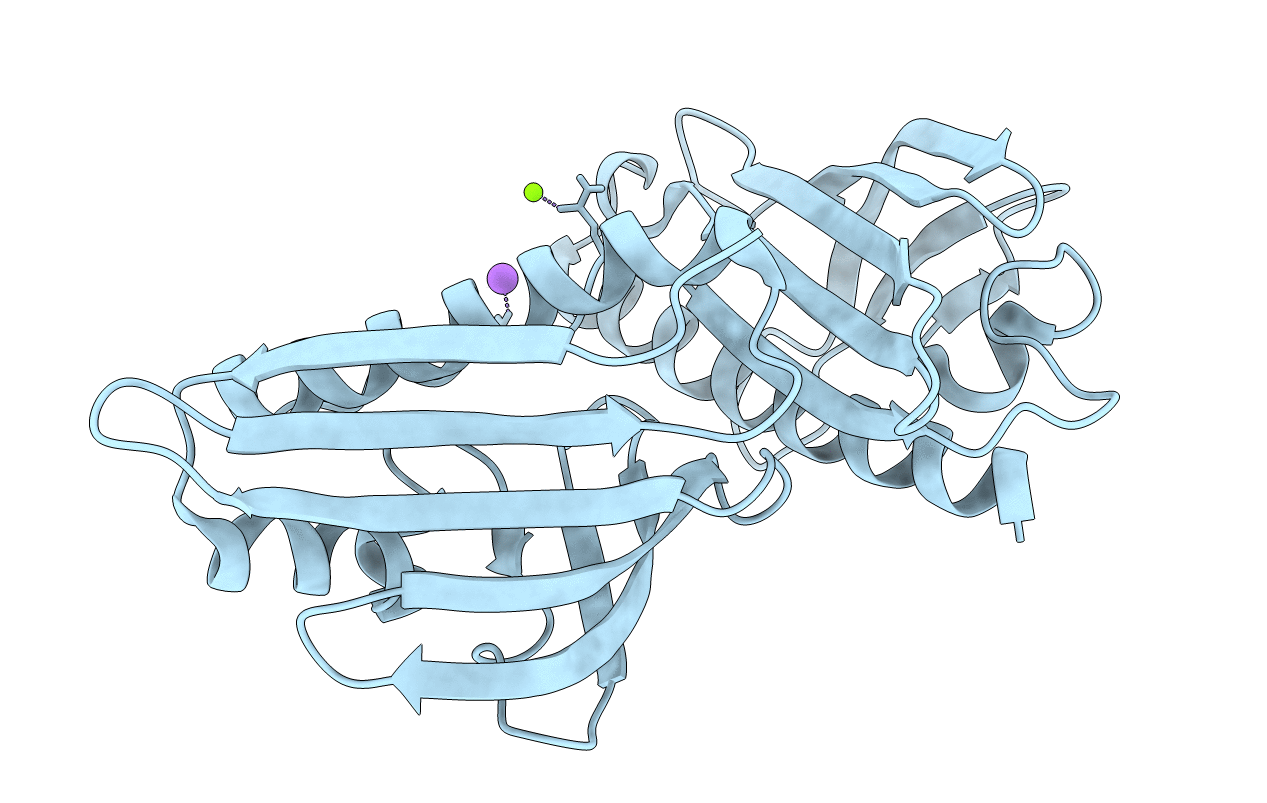 |
Dynu16 Crystal Structure, A Putative Protein In The Dynemicin Biosynthetic Locus
Organism: Micromonospora chersina
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-11-18 Classification: UNKNOWN FUNCTION |
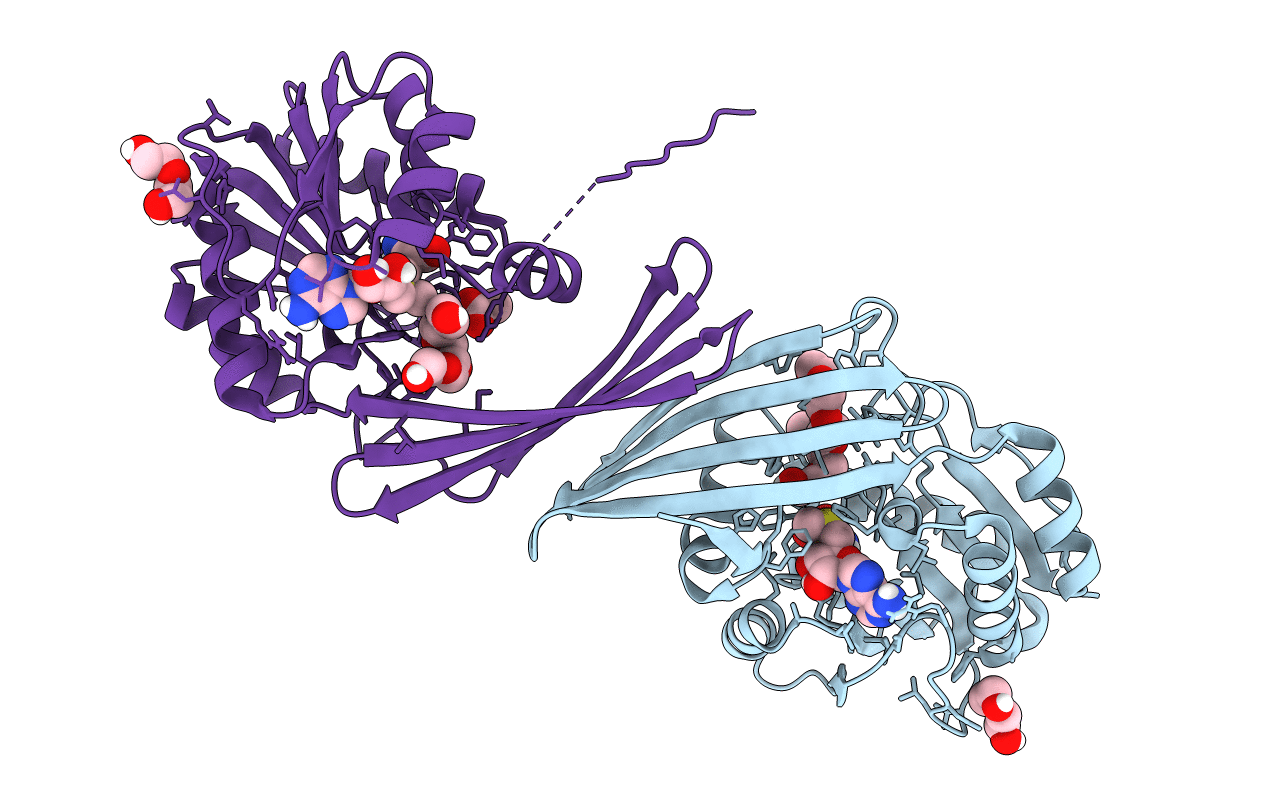 |
Structure Of Sam Bound Cals10, An Amino Pentose Methyltransferase From Micromonospora Echinaspora Involved In Calicheamicin Biosynthesis
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-10-07 Classification: TRANSFERASE Ligands: PEG, SAM, ACT, 1PE |
 |
Structure Of Dynf From The Dynemicin Biosynthesis Pathway Of Micromonospora Chersina
Organism: Micromonospora chersina
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-09-16 Classification: UNKNOWN FUNCTION Ligands: PLM |
 |
The X-Ray Crystal Structure Of Absh3, An Fad Dependent Reductase From The Abyssomicin Biosynthesis Pathway In Streptomyces
Organism: Streptomyces sp. lc-6-2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-11-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, CL |
 |
Crystal Structure Of Carminomycin-4-O-Methyltransferase Dnrk In Complex With Tetrazole-Sah
Organism: Streptomyces peucetius
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2015-12-23 Classification: TRANSFERASE Ligands: S8M |
 |
Crystal Structure Of Carminomycin-4-O-Methyltransferase Dnrk In Complex With Sah And 2-Chloro-4-Nitrophenol
Organism: Streptomyces peucetius
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2015-12-16 Classification: TRANSFERASE Ligands: SAH, P9P, SO4 |
 |
Organism: Streptomyces globisporus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-06-12 Classification: Lyase, Transferase |

