Search Count: 11
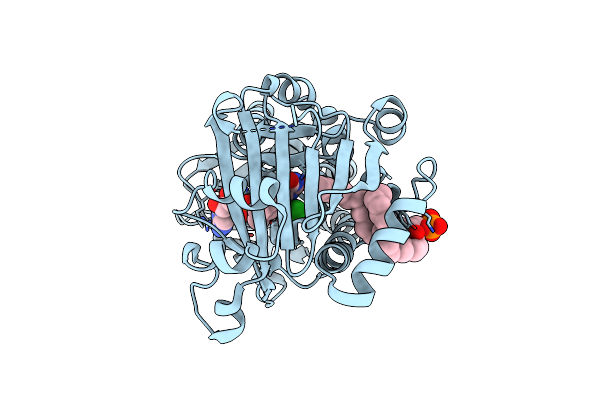 |
Crystal Structure Of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid- Assisted Flavoprotein Strategy For Regioselective Aromatic Hydroxylation: Platinum Derivative
Organism: Rhodococcus jostii
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2013-07-24 Classification: OXIDOREDUCTASE Ligands: FAD, P3A, CL |
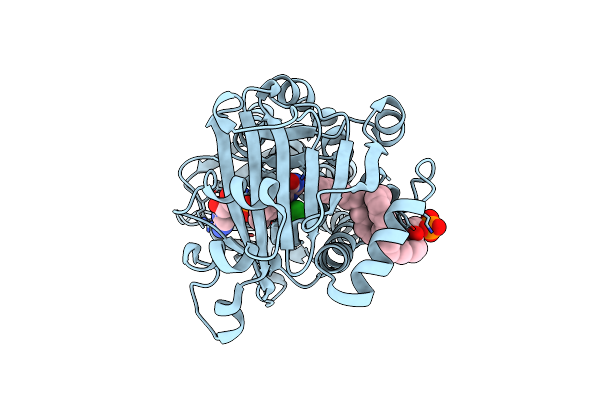 |
Crystal Structure Of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid- Assisted Flavoprotein Strategy For Regioselective Aromatic Hydroxylation: Native Data
Organism: Rhodococcus jostii
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2013-07-24 Classification: OXIDOREDUCTASE Ligands: FAD, P3A, CL |
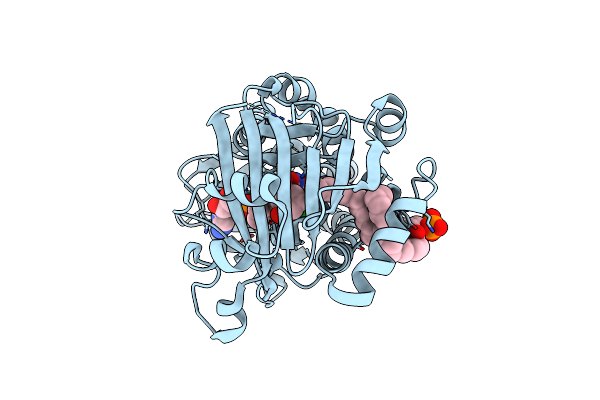 |
Crystal Structure Of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid- Assisted Flavoprotein Strategy For Regioselective Aromatic Hydroxylation: H213S Mutant In Complex With 3-Hydroxybenzoate
Organism: Rhodococcus jostii
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2013-07-24 Classification: OXIDOREDUCTASE Ligands: FAD, P3A, 3HB, CL |
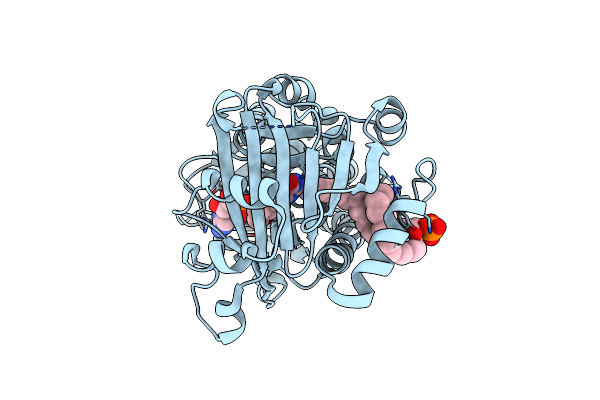 |
Crystal Structure Of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid- Assisted Flavoprotein Strategy For Regioselective Aromatic Hydroxylation: Q301E Mutant
Organism: Rhodococcus jostii
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2013-07-24 Classification: OXIDOREDUCTASE Ligands: FAD, P3A |
 |
Crystal Structure Of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid- Assisted Flavoprotein Strategy For Regioselective Aromatic Hydroxylation: Y105F Mutant
Organism: Rhodococcus jostii
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2013-07-24 Classification: OXIDOREDUCTASE Ligands: FAD, P3A, CL |
 |
Structure-Based Redesign Of Cofactor Binding In Putrescine Oxidase: Wild Type Enzyme
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-05-18 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, SO4 |
 |
Structure-Based Redesign Of Cofactor Binding In Putrescine Oxidase: Wild Type Bound To Putrescine
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-05-18 Classification: OXIDOREDUCTASE Ligands: FAD, 4HA, GOL, SO4 |
 |
Structure-Based Redesign Of Cofactor Binding In Putrescine Oxidase: A394C Mutant
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-05-18 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, GOL |
 |
Structure-Based Redesign Of Cofactor Binding In Putrescine Oxidase: P15I-A394C Double Mutant
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-05-18 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, GOL |
 |
Structure-Based Redesign Of Cofactor Binding In Putrescine Oxidase: A394C-A396T-Q431G Triple Mutant
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2011-05-18 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
The Crystal Structure Of Chlorite Dismutase: A Detox Enzyme Producing Molecular Oxygen
Organism: Azospira oryzae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-03-03 Classification: OXIDOREDUCTASE Ligands: HEM, SCN, CO3 |

