Search Count: 77
 |
Cryoem Structure Of Thermocas9 In Post-Cleavage State Bound With The Dna Containing Nnnncca Pam
Organism: Geobacillus thermodenitrificans, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: MG, GTP |
 |
Cryoem Structure Of Thermocas9 In Pre-Cleavage State Bound With The Dna Containing Nnnncca Pam
Organism: Geobacillus thermodenitrificans
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN/DNA/RNA |
 |
Organism: Geobacillus thermodenitrificans
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: GTP, MG |
 |
Organism: Haliangium ochraceum, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: IMMUNE SYSTEM |
 |
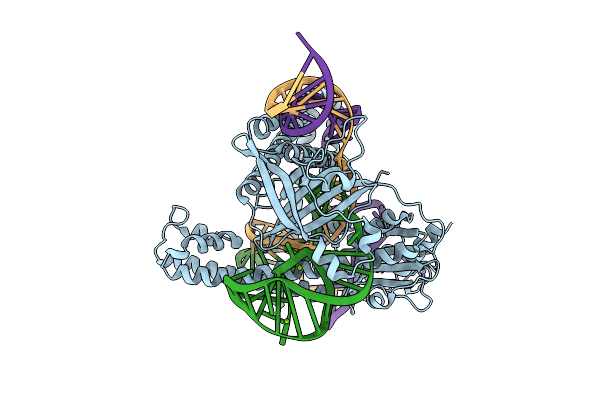
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
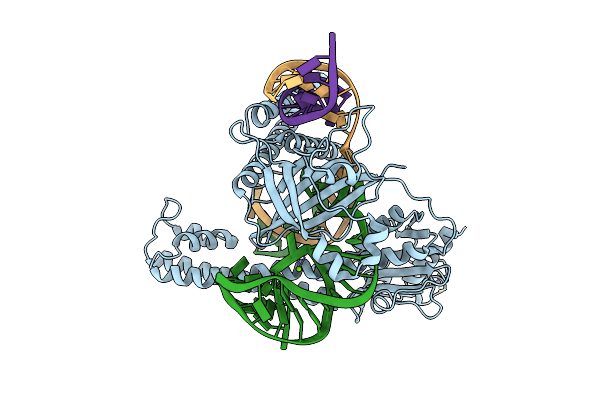
Organism: Mycolicibacterium mucogenicum
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: RNA BINDING PROTEIN Ligands: MG, ZN |
 |
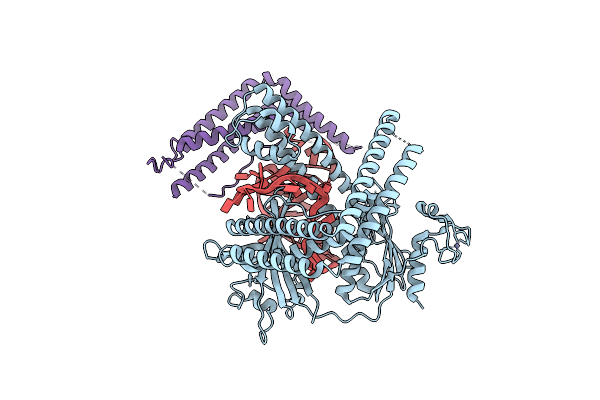
Cryo-Em Structure Of The Cas12M2-Crrna-Target Dna Ternary Complex Intermediate State
Organism: Mycolicibacterium mucogenicum
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: RNA BINDING PROTEIN Ligands: MG, ZN |
 |
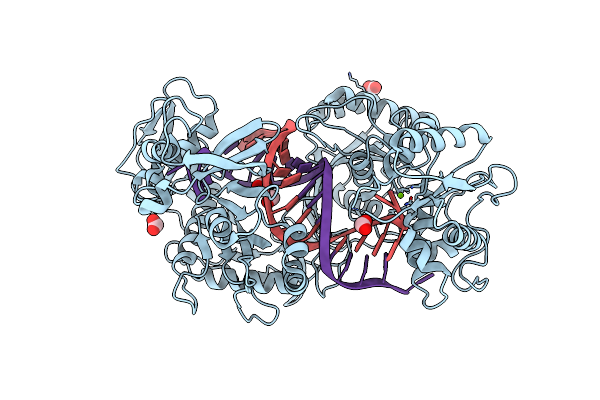
Organism: Mycolicibacterium mucogenicum
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: RNA BINDING PROTEIN Ligands: MG, ZN |
 |
Structure Of Clostridium Butyricum Argonaute Bound To A Guide Dna (5' Deoxycytidine) And A 19-Mer Target Dna
Organism: Clostridium butyricum, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.55 Å Release Date: 2019-04-24 Classification: HYDROLASE Ligands: MG, FMT |
 |
Crystal Structure Of Catalytically Inactive Fncas12 Mutant Bound To An R-Loop Structure Containing A Pre-Crrna Mimic And Full-Length Dna Target
Organism: Francisella tularensis subsp. novicida (strain u112), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: MG, B3P |
 |
Organism: Francisella tularensis subsp. novicida (strain u112), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.34 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: MG |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-05-25 Classification: HYDROLASE Ligands: COA, CL, PGE, FLC, PEG |
 |
Organism: Thermobaculum terrenum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-11-19 Classification: HYDROLASE Ligands: NI |
 |
Crystal Structure Of Crispr-Associated Protein In Complex With 2'-Deoxyadenosine 5'-Triphosphate
Organism: Thermobaculum terrenum
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2014-11-19 Classification: HYDROLASE Ligands: NI, MG, DTP |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-10-15 Classification: DNA BINDING PROTEIN Ligands: CA |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-10-01 Classification: HYDROLASE/DNA |
 |
Crystal Structure Of The E. Coli Crispr Rna-Guided Surveillance Complex, Cascade
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2014-08-13 Classification: Hydrolase/RNA Ligands: ZN |
 |
The Crystal Structure Of The P132R, Y133G Mutant Of Pyrococcus Furiosus Phosphoglucose Isomerase In Complex With Manganese And 5-Phospho-D-Arabinonate.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2014-07-30 Classification: ISOMERASE Ligands: MN, PA5, EDO |
 |
The Crystal Structure Of The P132A, Y133G Mutant Of Pyrococcus Furiosus Phosphoglucose Isomerase In Complex With Manganese And 5-Phospho-D-Arabinonate.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2014-07-30 Classification: ISOMERASE Ligands: MN, PA5, EDO |
 |
The Crystal Structure Of The P132A, Y133D Mutant Of Pyrococcus Furiosus Phosphoglucose Isomerase In Complex With Manganese.
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2014-07-30 Classification: ISOMERASE Ligands: MN |

