Search Count: 20
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: DEP |
 |
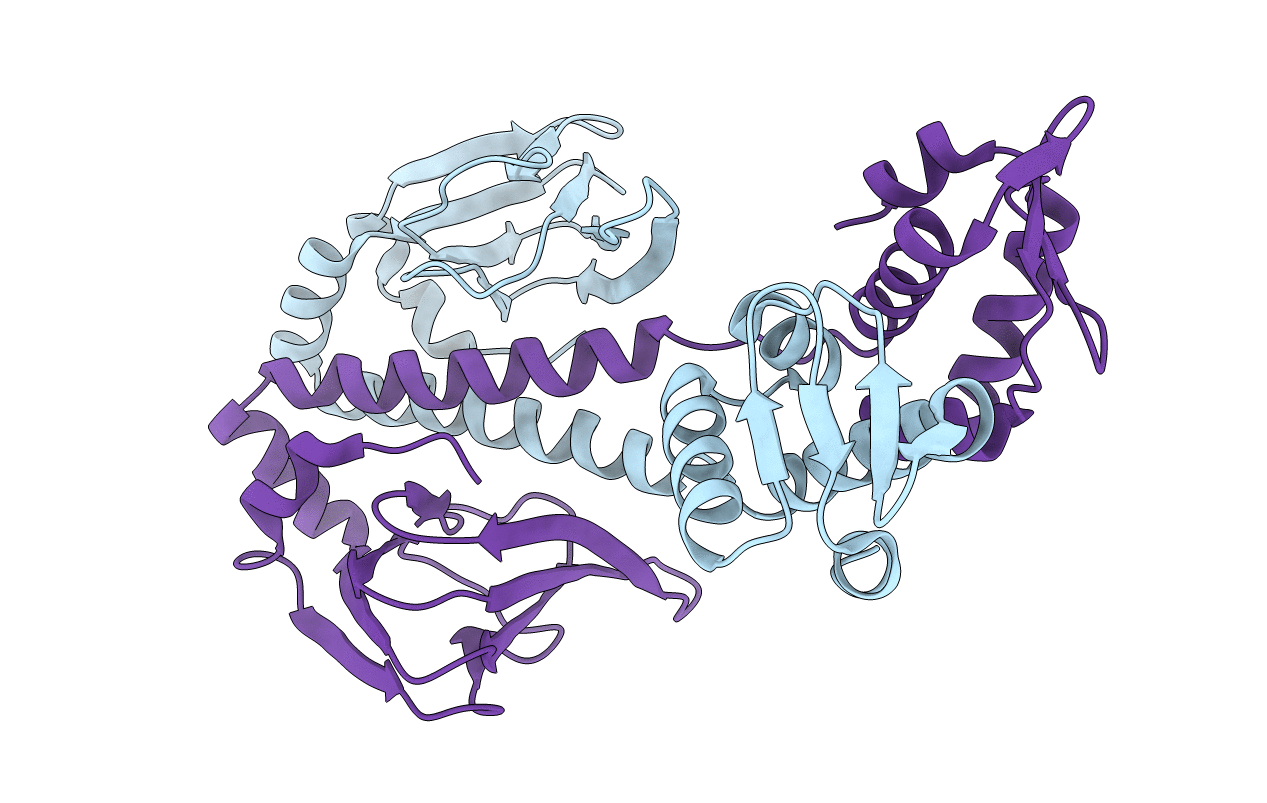
Crystal Structure Of Oxidized Cprk In Complex With O-Chlorophenolacetic Acid
Organism: Desulfitobacterium hafniense
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-07-04 Classification: DNA BINDING PROTEIN Ligands: SO4, 3C4 |
 |
Organism: Desulfitobacterium dehalogenans
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2006-07-04 Classification: DNA BINDING PROTEIN |
 |
Nmr-Based Model Of The Complex Of The Toxin Kid And A 5-Nucleotide Substrate Rna Fragment (Auaca)
|
 |
Structure Of The Octameric Flavoenzyme Vanillyl-Alcohol Oxidase: The505Ser Mutant
Organism: Penicillium simplicissimum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-07-02 Classification: OXIDOREDUCTASE Ligands: FAD, EUG |
 |
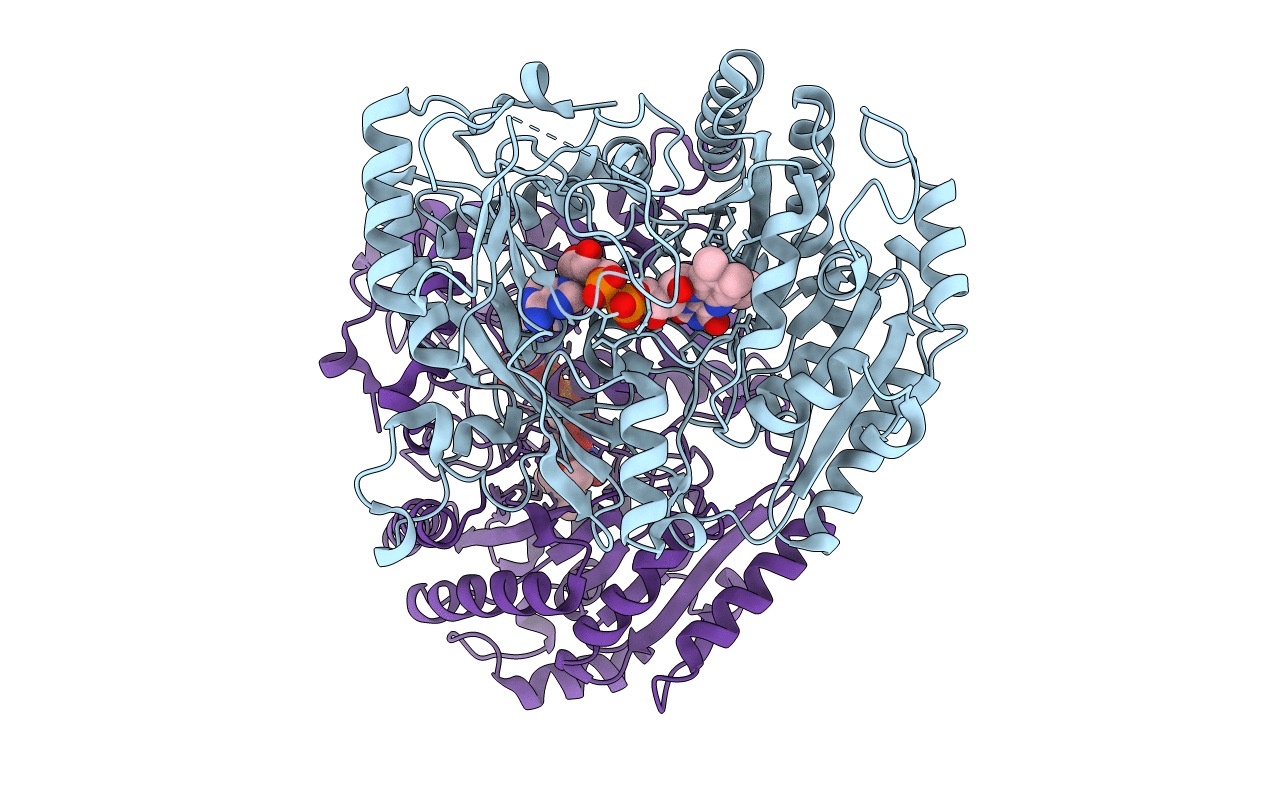
Structure Of The Octameric Flavoenzyme Vanillyl-Alcohol Oxidase: Ile238Thr Mutant
Organism: Penicillium simplicissimum
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2004-07-02 Classification: OXIDOREDUCTASE Ligands: FAD, EUG |
 |
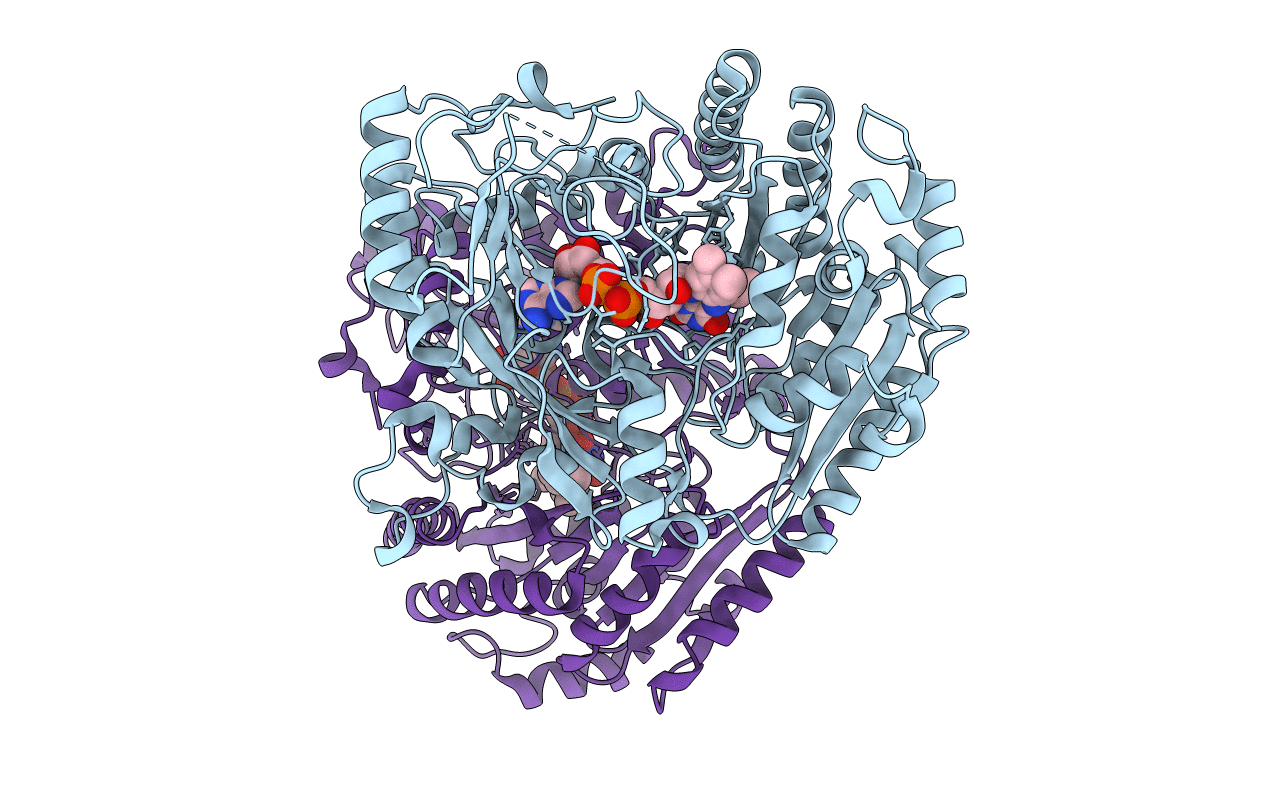
Structure Of The Octameric Flavoenzyme Vanillyl-Alcohol Oxidase: Phe454Tyr Mutant
Organism: Penicillium simplicissimum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-07-02 Classification: OXIDOREDUCTASE Ligands: FAD, EUG |
 |
Structure Of The Octameric Flavoenzyme Vanillyl-Alcohol Oxidase: Glu502Gly Mutant
Organism: Penicillium simplicissimum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2004-07-02 Classification: OXIDOREDUCTASE Ligands: FAD, EUG |
 |
Organism: Geobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-04-06 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
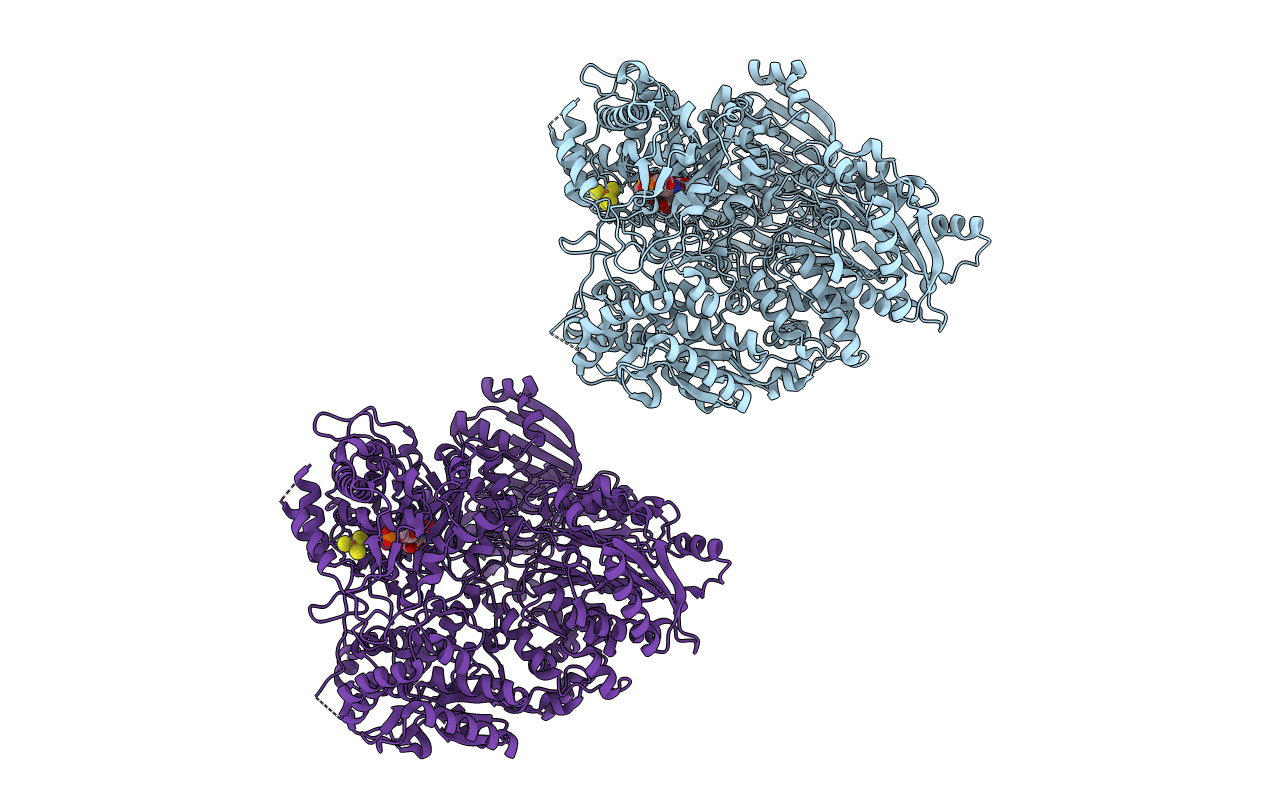
Organism: Geobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-04-06 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
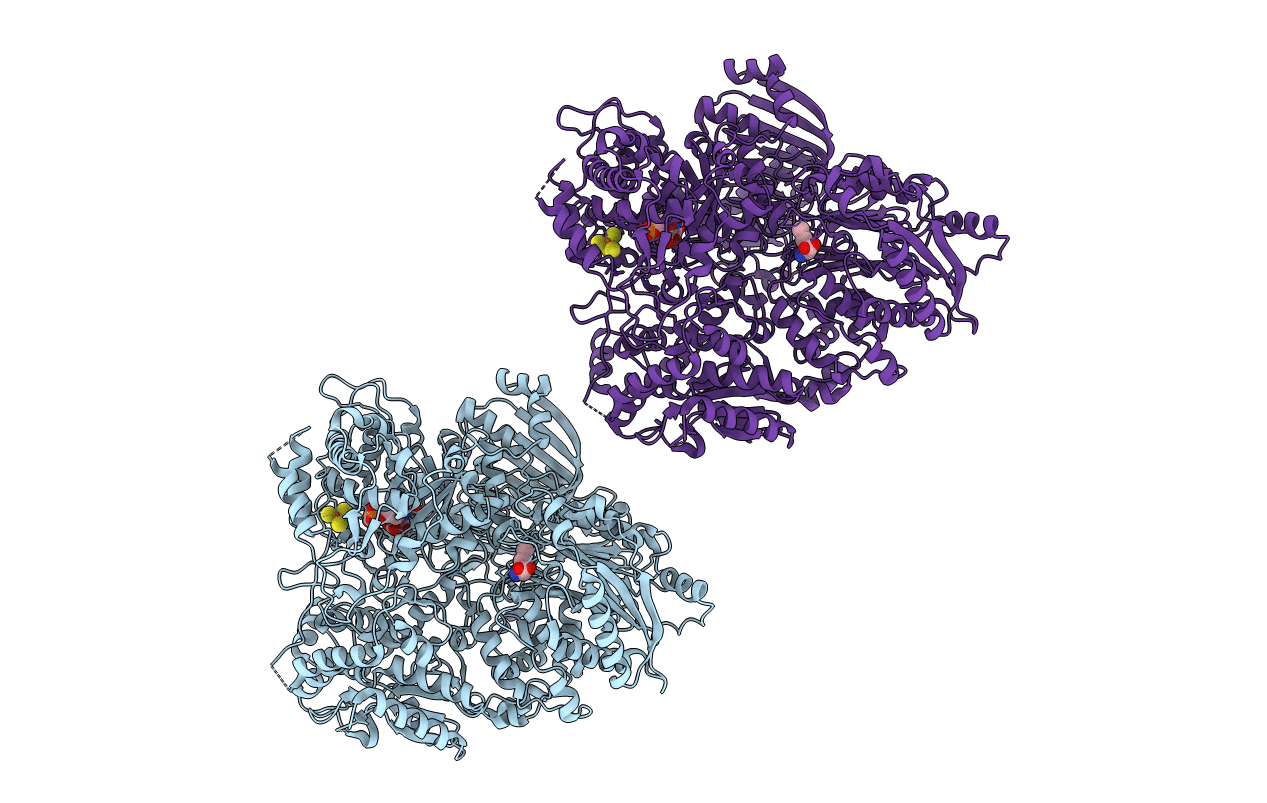
Glutamate Synthase From Synechocystis Sp In Complex With 2-Oxoglutarate At 2.0 Angstrom Resolution
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-06-11 Classification: OXIDOREDUCTASE Ligands: FMN, F3S, AKG |
 |
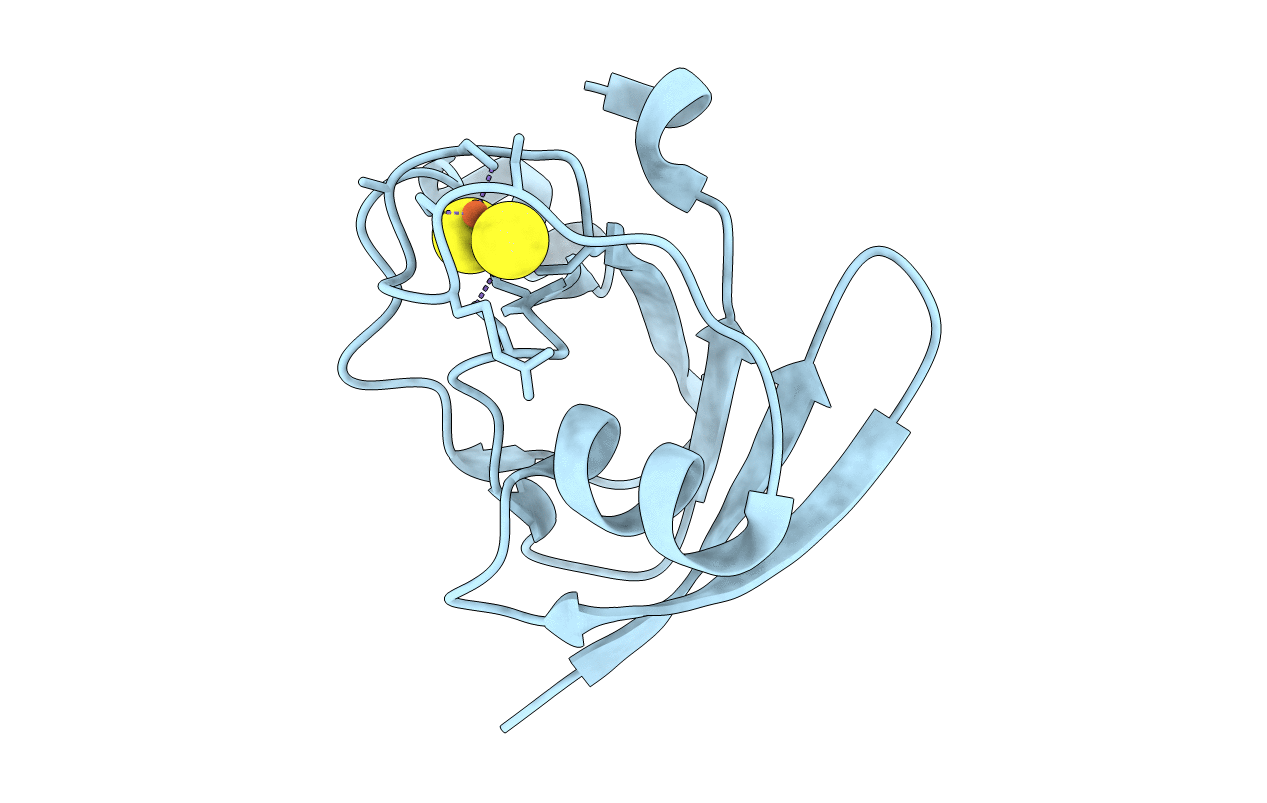
Glutamate Synthase From Synechocystis Sp In Complex With 2-Oxoglutarate And L-Don At 2.45 Angstrom Resolution
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2003-06-11 Classification: OXIDOREDUCTASE Ligands: FMN, F3S, AKG, ONL |
 |
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-06-11 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
Structural Studies On The Synchronization Of Catalytic Centers In Glutamate Synthase: Complex With 2-Oxoglutarate
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2002-07-31 Classification: OXIDOREDUCTASE Ligands: FMN, F3S, AKG |
 |
Structural Studies On The Synchronization Of Catalytic Centers In Glutamate Synthase: Reduced Enzyme
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2002-07-31 Classification: OXIDOREDUCTASE Ligands: FMN, F3S |
 |
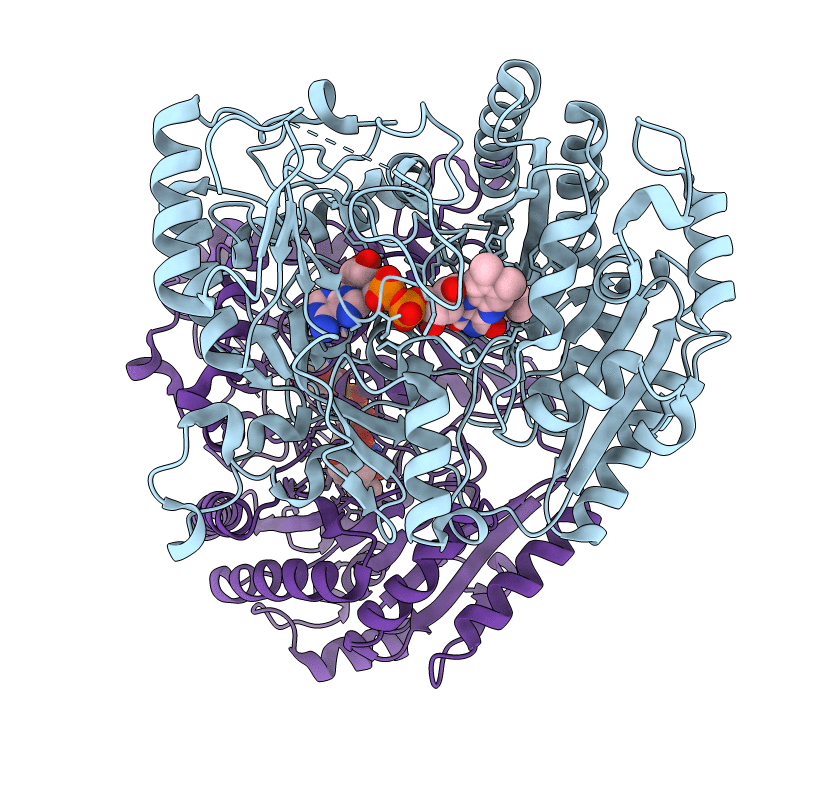
Structural Studies On The Synchronization Of Catalytic Centers In Glutamate Synthase: Native Enzyme
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2002-07-31 Classification: OXIDOREDUCTASE Ligands: ACT, FMN, F3S |
 |
Organism: Penicillium simplicissimum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2000-03-07 Classification: FLAVIN-DEPENDENT OXIDASE ENZYME Ligands: FAD, EUG |
 |
Organism: Penicillium simplicissimum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1999-09-20 Classification: FLAVOENZYME Ligands: FAD, ACT |
 |
Structure Of The H422A Mutant Vanillyl-Alcohol Oxidase In Complex With Isoeugenol
Organism: Penicillium simplicissimum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1999-09-20 Classification: OXIDOREDUCTASE Ligands: FAD, EUG |

