Search Count: 40
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SEY, PO4, CL |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SIN, EDO, PO4, CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-20 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Clostridium carboxidivorans p7
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-03-13 Classification: TRANSPORT PROTEIN Ligands: CL |
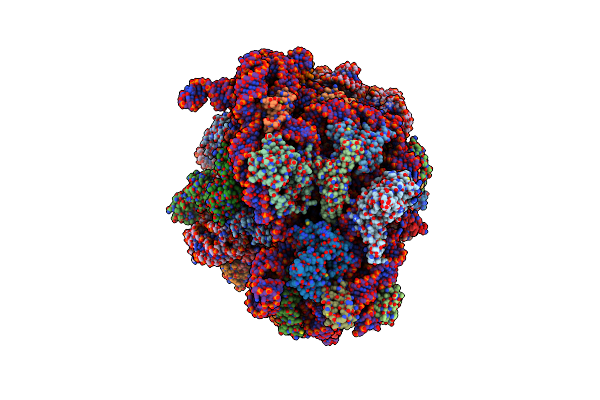 |
Cryo-Em Structure Of P. Urativorans 70S Ribosome In Complex With Hibernation Factors Balon And Raia (Structure 1).
Organism: Psychrobacter urativorans
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: RIBOSOME Ligands: MG |
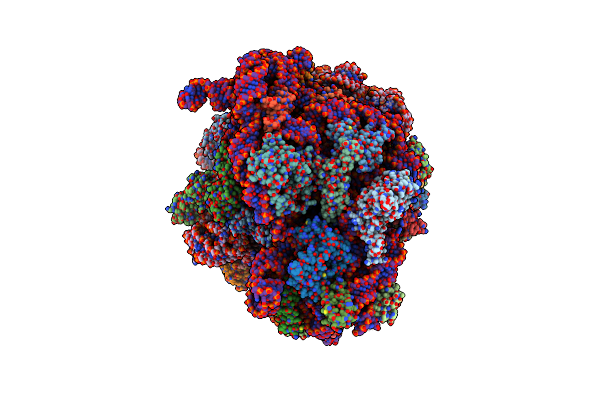 |
Cryo-Em Structure Of P. Urativorans 70S Ribosome In Complex With Hibernation Factor Balon, Mrna And P-Site Trna (Structure 2).
Organism: Psychrobacter urativorans
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: RIBOSOME |
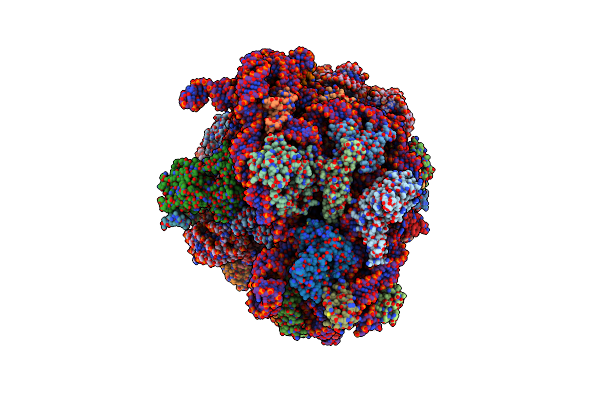 |
Cryo-Em Structure Of P. Urativorans 70S Ribosome In Complex With Hibernation Factor Balon And Ef-Tu(Gdp) (Structure 3).
Organism: Psychrobacter urativorans
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: RIBOSOME Ligands: GDP, MG |
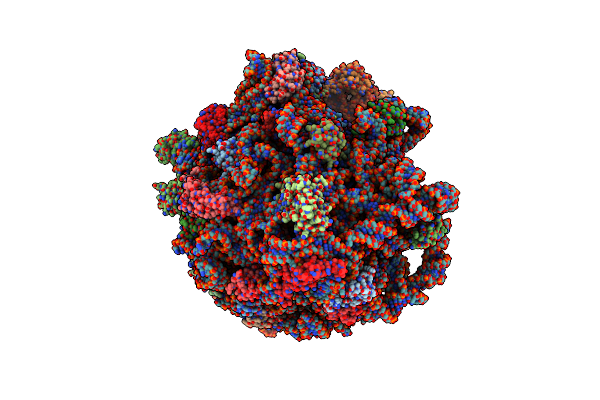 |
Cryo-Em Structure Of The Mycobacterium Smegmatis 70S Ribosome In Complex With Hibernation Factor Msmeg1130 (Balon) (Structure 4)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: ZN |
 |
Cryo-Em Structure Of The Mycobacterium Smegmatis 70S Ribosome In Complex With Hibernation Factor Rv2629 (Balon) (Structure 5)
Organism: Mycobacterium tuberculosis h37rv, Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: ZN |
 |
Cryo-Em Structure Of The Mycobacterium Smegmatis 70S Ribosome In Complex With Hibernation Factor Msmeg1130 (Balon) And Msmegef-Tu(Gdp) (Composite Structure 6)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: GDP, ZN |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul1 In Complex With 6-Hydroxymethylpterin
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR, GOL, SO4, CL |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: SO4, CL, GOL, PEG |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul2 In Complex With 7,8-Dihydropteroate, Magnesium, And Pyrophosphate
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: 78H, POP, MG, GOL, CL, PAB |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: GOL, CL, SO4 |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul3 In Complex With 6-Hydroxymethylpterin
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR |
 |
Crystal Structure Of Adaptive Laboratory Evolved Sulfonamide-Resistant Dihydropteroate Synthase (Dhps) From Escherichia Coli In Complex With 6-Hydroxymethylpterin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul3 In Complex With Reaction Intermediate
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: XHP, PAB, GOL, CL, MG, POP |
 |
Crystal Structure Of Autotaxin (Enpp2) With 18F-Labeled Positron Emission Tomography Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: NAG, SCN, ZN, CA, IOD, 4I0 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: NAP, MG |

