Search Count: 112
 |
Her3 Receptor In Complex With The Fab Fragment Of Tk-Hu A3 Monoclonal Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: ANTITUMOR PROTEIN Ligands: NAG, SCN |
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus Bound To Sinapic Acid, Tetragonal Crystal
Organism: Trametes cinnabarina
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: FAD, SXX, SO4, NAG |
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus Bound To Sinapic Acid, Orthorhombic Crystal
Organism: Trametes cinnabarina
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: FAD, SXX, NAG, SO4 |
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus Bound To Guaiacol, Orthorhombic Crystal
Organism: Trametes cinnabarina
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, SO4, JZ3 |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: HEM, TES, FMT, NA |
 |
Olep In Complex With Lithocholic Acid In High Salt Crystallization Conditions
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: HEM, 4OA, FMT, NA |
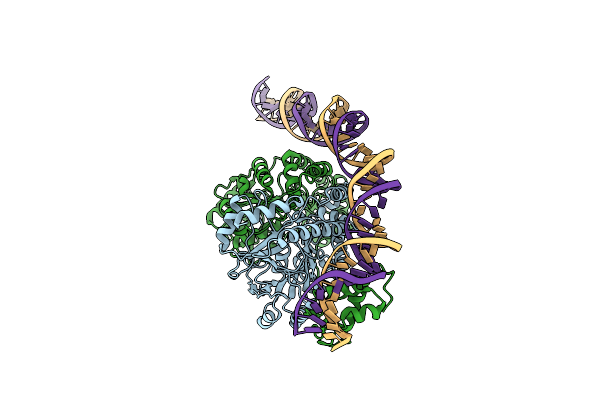 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Half-Closed Conformation
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
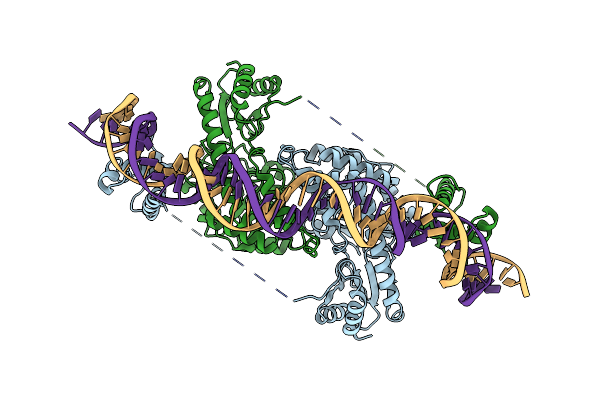 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Closed Conformation, C2 Symmetry.
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
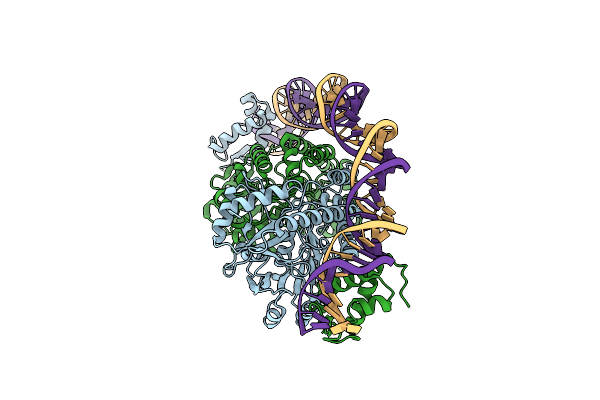 |
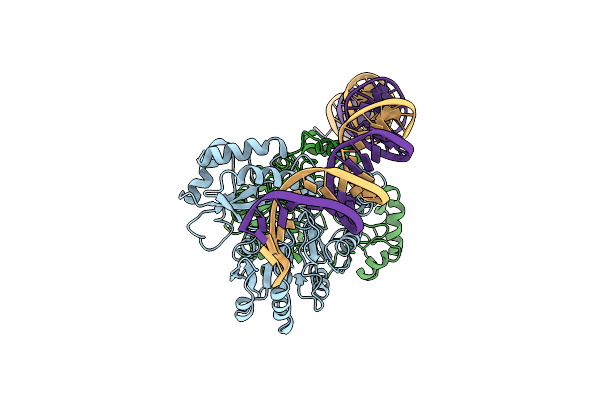
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Closed Conformation, C1 Symmetry
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Open Conformation
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
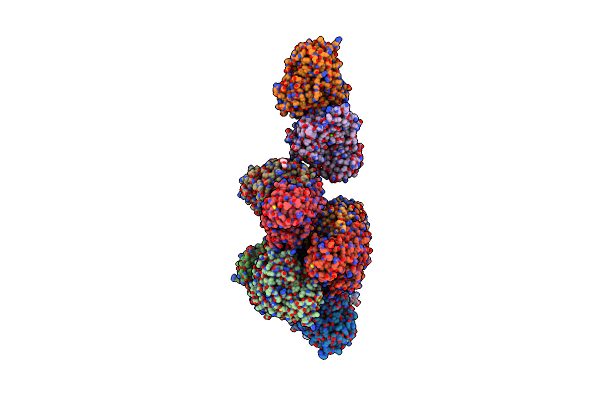 |
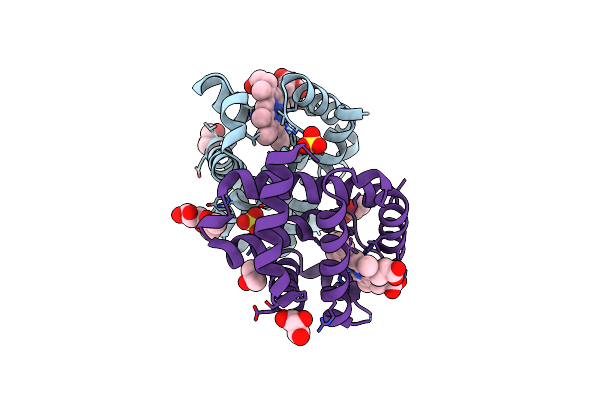
Organism: Alkalihalobacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-09-28 Classification: DNA BINDING PROTEIN Ligands: CA, CL, PGE, EDO, PEG, P6G |
 |
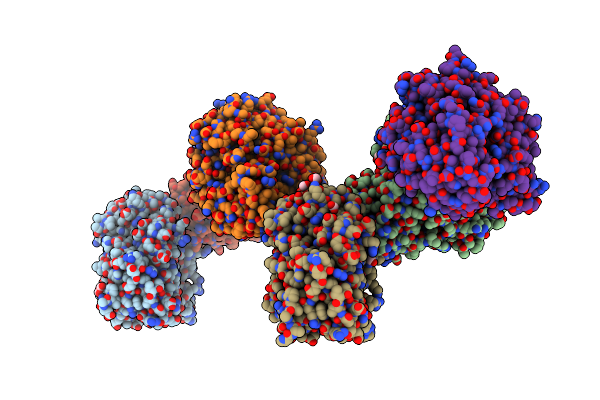
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-07-13 Classification: OXYGEN TRANSPORT Ligands: HEM, SO4, GOL, IPA, BTB |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2022-01-26 Classification: OXIDOREDUCTASE Ligands: HEM, DEB, GOL, FMT |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-01-26 Classification: OXIDOREDUCTASE Ligands: HEM, DEB, FMT |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-01-26 Classification: OXIDOREDUCTASE Ligands: HEM, DEB, FMT, NA, GOL |
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus, Ligand-Free Form
Organism: Pycnoporus cinnabarinus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-02-03 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, SO4 |
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus, Glucose-Bound Form
Organism: Pycnoporus cinnabarinus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-02-03 Classification: OXIDOREDUCTASE Ligands: FAD, BGC, GLC, NAG, SO4 |
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus, Laminaribiose-Bound Form
Organism: Trametes cinnabarina
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-02-03 Classification: OXIDOREDUCTASE Ligands: FAD, GLC, NAG, SO4 |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-10-21 Classification: OXIDOREDUCTASE Ligands: HEM, QR8, TRS, FMT, GOL, NA |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Release Date: 2020-10-21 Classification: OXIDOREDUCTASE Ligands: HEM, QR8 |

