Search Count: 52
 |
Co-Crystal Of Broadly Neutralizing Biparatopic Vhh In Complex With Cardiotoxin (P01468) Naja Pallida
Organism: Vicugna pacos, Naja pallida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TOXIN Ligands: ACT |
 |
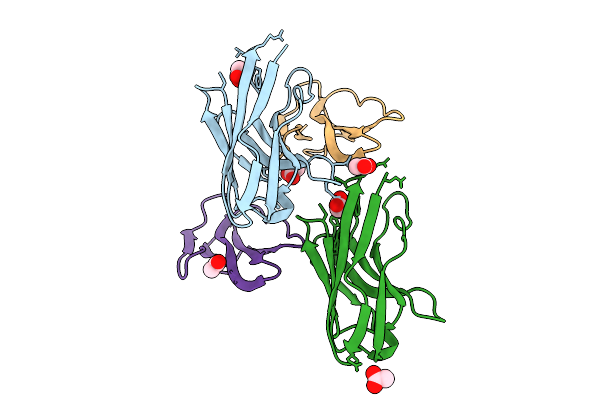
Co-Crystal Of Broadly Neutralizing Vhh In Complex With Short Neurotoxin 1 (P01426) Naja Pallida
Organism: Vicugna pacos, Naja pallida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TOXIN Ligands: NI |
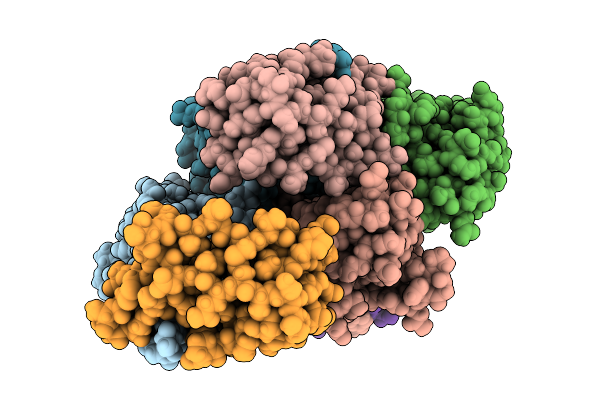 |
Organism: Vicugna pacos, Naja kaouthia
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: TOXIN Ligands: HOH |
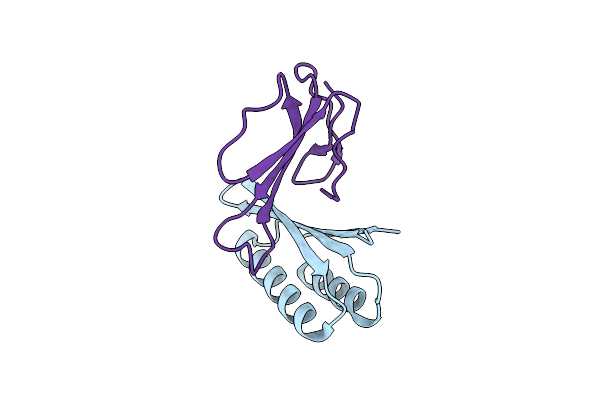 |
Organism: Synthetic construct, Naja kaouthia
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2024-10-09 Classification: DE NOVO PROTEIN/TOXIN |
 |
Organism: Synthetic construct, Naja pallida
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-09 Classification: DE NOVO PROTEIN/TOXIN |
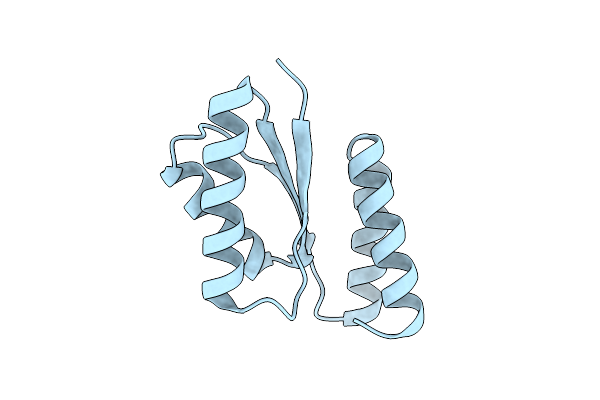 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-10-09 Classification: DE NOVO PROTEIN |
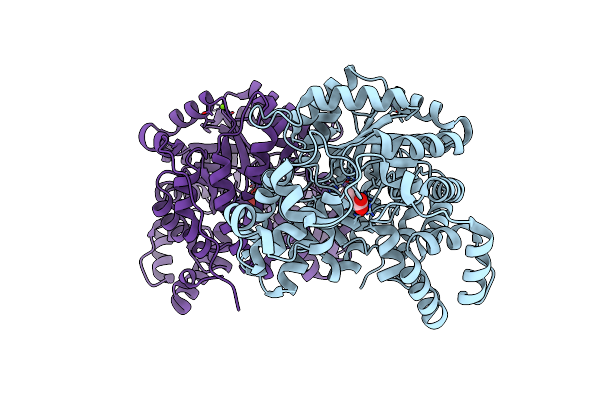 |
Cryo-Em Structure Of "Ct-Ct Dimer" Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-12-28 Classification: LIGASE Ligands: MG, MN, PYR |
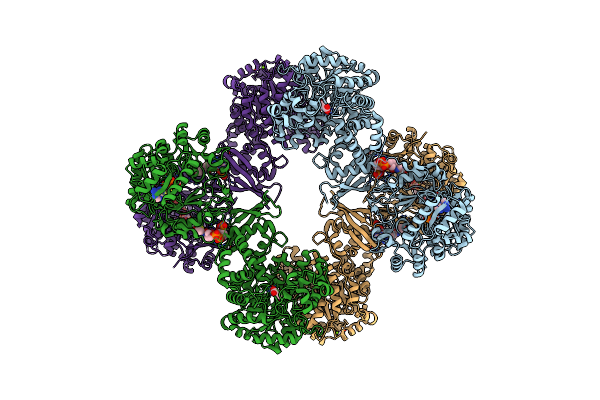 |
Cryo-Em Structure Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: MG, MN, ADP, ACO, PYR, BCT |
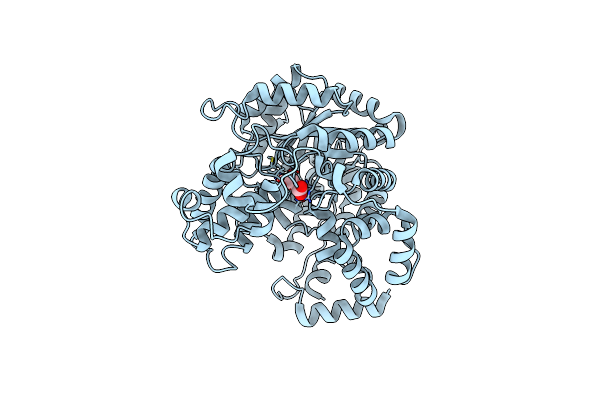 |
Cryo-Em Structure Of "Ct Oxa" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: MN, OAA |
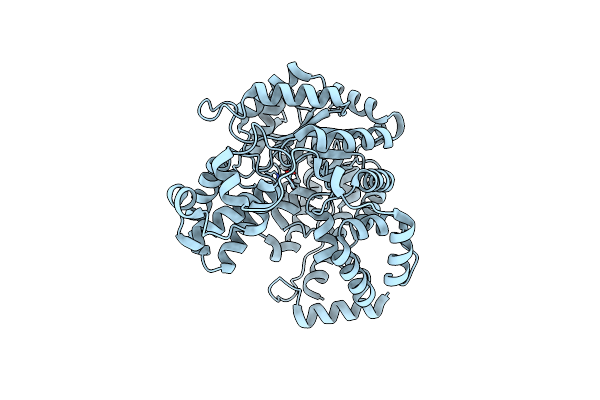 |
Cryo-Em Structure Of "Ct Empty" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: MG, MN |
 |
Cryo-Em Structure Of "Ct React" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: MG, MN, OAA, BTN |
 |
Cryo-Em Structure Of "Ct Pyr" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: MG, MN, PYR |
 |
Cryo-Em Structure Of "Bc React" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: BTN, MG, MN, ATP, ACO, PYR, BCT, ADP |
 |
Cryo-Em Structure Of "Bc Closed" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: ATP, MG, ACO |
 |
Cryo-Em Structure Of "Bc Open" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: ACO, BCT |
 |
Cryo-Em Structure Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa And Cyclic Di-Amp
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: MG, MN, ADP, ACO, PYR, 2BA |
 |
Organism: Escherichia coli, Escherichia coli 4.0522
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: RIBOSOME Ligands: MG, ZN |

