Search Count: 30
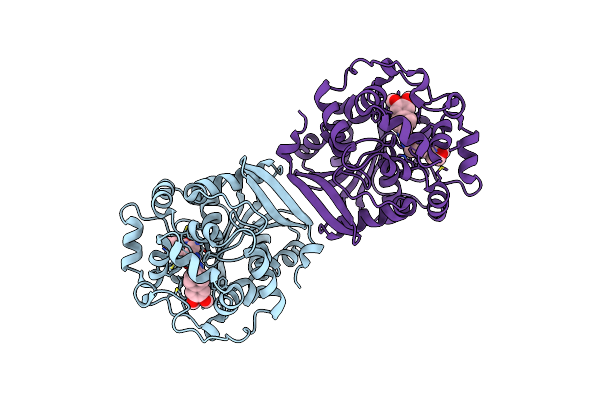 |
Crystal Structure Of Human Soluble Epoxide Hydrolase C-Terminal Domain In Complex With A Benzohomoadamantane-Based Urea Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: HYDROLASE Ligands: A1H8Y |
 |
Organism: Enterococcus italicus dsm 15952, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: RNA BINDING PROTEIN |
 |
Organism: Enterococcus italicus dsm 15952, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: RNA BINDING PROTEIN |
 |
Organism: Enterococcus italicus dsm 15952
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: RNA BINDING PROTEIN |
 |
Organism: Enterococcus italicus dsm 15952
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: RNA BINDING PROTEIN |
 |
Cryoem Structure Of Enterococcus Italicus Csm-Crrna Complex Bound To Ampnpp
Organism: Enterococcus italicus dsm 15952
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: RNA BINDING PROTEIN Ligands: ZAN, MG |
 |
Cryoem Structure Of Enterococcus Italicus Csm-Crrna-Ctr Complex Bound To Ampnpp
Organism: Enterococcus italicus dsm 15952, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: RNA BINDING PROTEIN Ligands: ZAN, MG |
 |
Cryoem Structure Of Enterococcus Italicus Csm-Crrna-Ctr1 Complex (4.3) Bound To Ampnpp
Organism: Enterococcus italicus dsm 15952, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: RNA BINDING PROTEIN Ligands: ZAN, MG |
 |
Cryoem Structure Of Enterococcus Italicus Csm-Crrna-Ctr1 Complex Bound To Pnppa3 And Ampnpp
Organism: Enterococcus italicus dsm 15952, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: RNA BINDING PROTEIN Ligands: A1II9, ANP, MG |
 |
Cryoem Structure Of Enterococcus Italicus Csm-Crrna-Ctr2 Complex Bound To Pnppa3 And Ampnpp
Organism: Enterococcus italicus dsm 15952, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: RNA BINDING PROTEIN Ligands: A1II9, ANP, MG |
 |
Cryoem Structure Of Enterococcus Italicus Csm-Crrna Complex Bound To Pnppa3 And Ampnpp
Organism: Enterococcus italicus dsm 15952
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: RNA BINDING PROTEIN Ligands: A1II9, ANP, MN |
 |
Cryoem Structure Of Enterococcus Italicus Csm-Crrna-Ctr2 Complex (4.3) Bound To Ampnpp
Organism: Enterococcus italicus dsm 15952, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: RNA BINDING PROTEIN Ligands: ZAN, MG |
 |
Organism: Serratia
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-10-26 Classification: RNA BINDING PROTEIN Ligands: CA, ACT |
 |
Organism: Serratia
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-10-26 Classification: RNA BINDING PROTEIN |
 |
Organism: Enterococcus italicus (strain dsm 15952 / lmg 22039 / tp 1.5)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2020-02-26 Classification: RNA BINDING PROTEIN Ligands: AF2, SO4 |
 |
Structure Of The Carboxy-Terminal Domain Of The Bacteriophage T5 L- Shaped Tail Fibre
Organism: Escherichia phage t5
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-12-16 Classification: VIRAL PROTEIN |
 |
Structure Of The Carboxy-Terminal Domain Of The Bacteriophage T5 L- Shaped Tail Fiber Without Its Intra-Molecular Chaperone Domain
Organism: Enterobacteria phage t5
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2015-08-05 Classification: VIRAL PROTEIN Ligands: GOL |
 |
Structure Of The Carboxy-Terminal Domain Of The Bacteriophage T5 L- Shaped Tail Fiber With Its Intra-Molecular Chaperone Domain
Organism: Enterobacteria phage t5
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2015-08-05 Classification: VIRAL PROTEIN Ligands: FLC |
 |
Native Structure Of The Lytic Chapk Domain Of The Endolysin Lysk From Staphylococcus Aureus Bacteriophage K
Organism: Staphylococcus phage k
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2014-08-06 Classification: HYDROLASE Ligands: GOL, CA, MES, NA, ZN |
 |
Methylmercury Chloride Derivative Structure Of The Lytic Chapk Domain Of The Endolysin Lysk From Staphylococcus Aureus Bacteriophage K
Organism: Kayvirus kay
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2014-08-06 Classification: VIRAL PROTEIN Ligands: CL, GOL, EPE, CA, MMC, HG |

