Search Count: 6
 |
The 1.67 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, OH, MG, VK3, F3S |
 |
The 1.52 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: ELECTRON TRANSPORT Ligands: O, MG, FCO, 3NI, VK3, F3S |
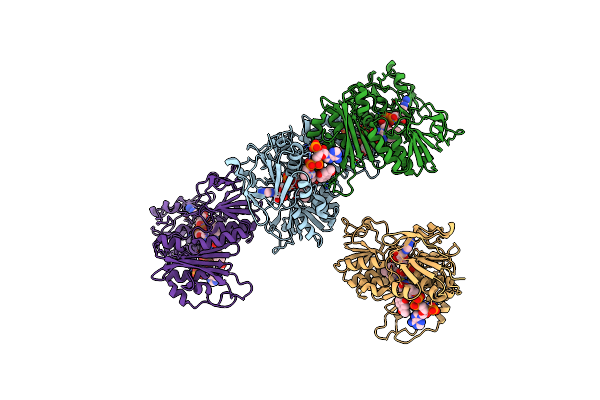 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-09-25 Classification: OXIDOREDUCTASE Ligands: FAD, COA, VK3 |
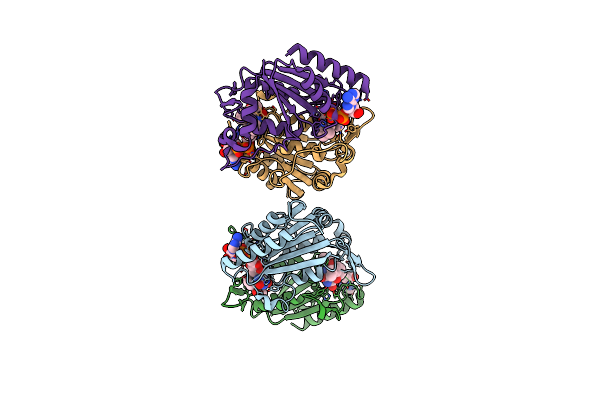 |
Complex Structure Of Nadph:Quinone Oxidoreductase With Menadione In Streptococcus Mutans
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-06-06 Classification: OXIDOREDUCTASE Ligands: FAD, VK3 |
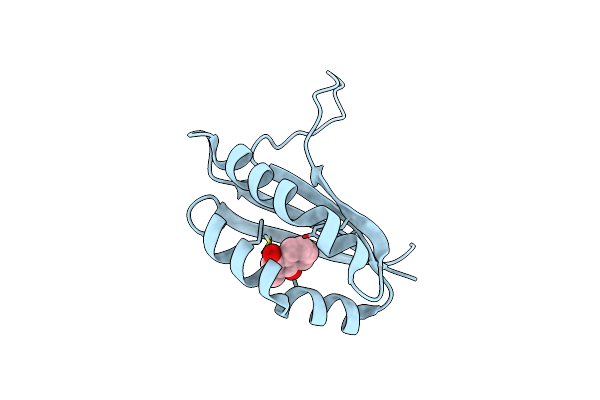 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2005-01-11 Classification: UNKNOWN FUNCTION Ligands: VK3 |
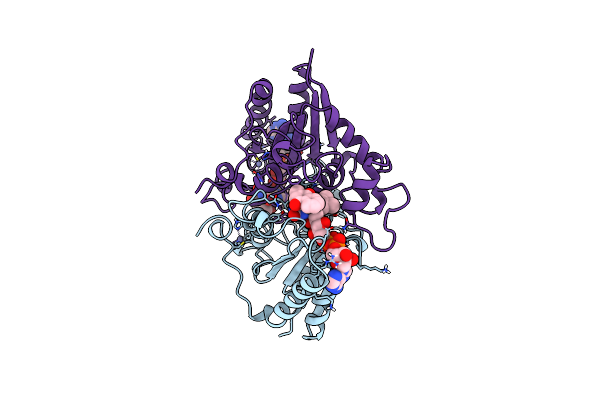 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 1999-08-18 Classification: OXIDOREDUCTASE Ligands: ZN, FAD, VK3 |

