Search Count: 355
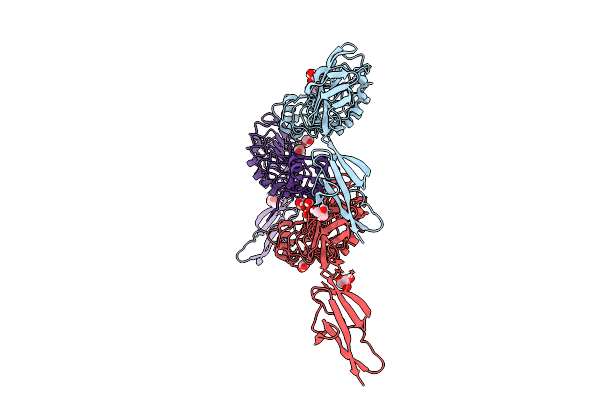 |
Inlb392_T336Y: T336Y Variant Of Listeria Monocytogenes Inlb (Internalin B) Residues 36-392
Organism: Listeria monocytogenes egd-e
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: CELL INVASION Ligands: GOL |
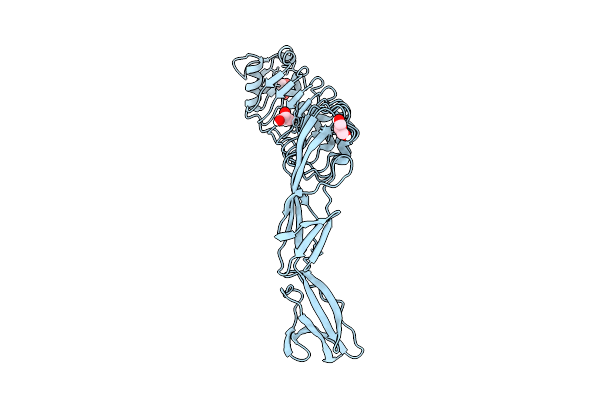 |
Inlb392_V333E: V333E Variant Of Listeria Monocytogenes Inlb (Internalin B) Residues 36-392
Organism: Listeria monocytogenes egd-e
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: CELL INVASION Ligands: GOL |
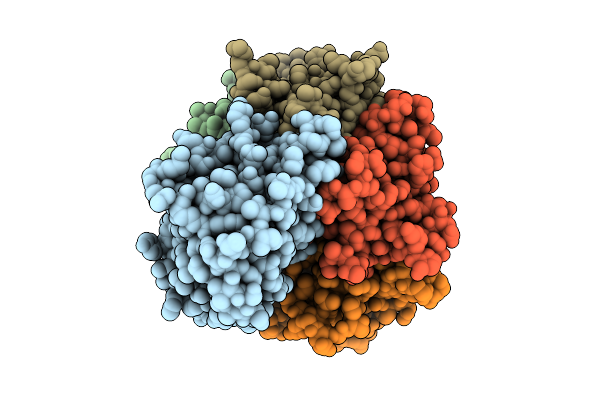 |
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN |
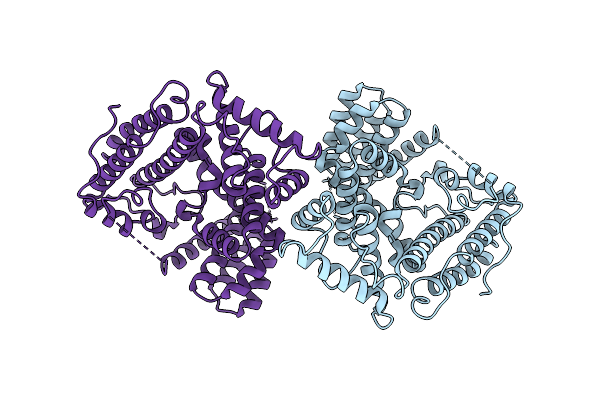 |
Organism: Puccinia graminis f. sp. tritici
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: ANTIFUNGAL PROTEIN |
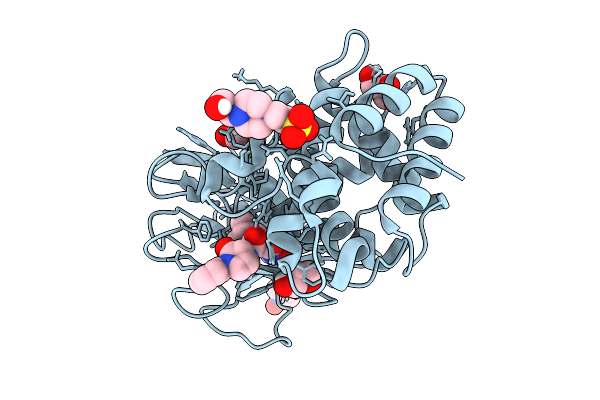 |
Crystal Structure Of Elastase Lasb From Pseudomonas Aeruginosa Pa14 In Complex With 6558
Organism: Pseudomonas aeruginosa pa14
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: HYDROLASE Ligands: ZN, CA, A1IM0, GOL, EPE |
 |
Organism: Brucella abortus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: GOL |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: ZN, ARG |
 |
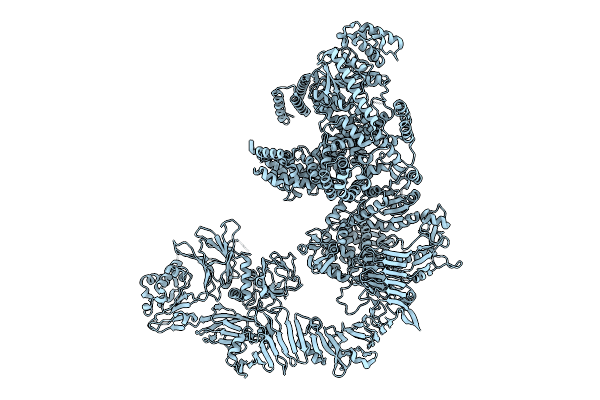
Organism: Staphylococcus aureus, Mammalia
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
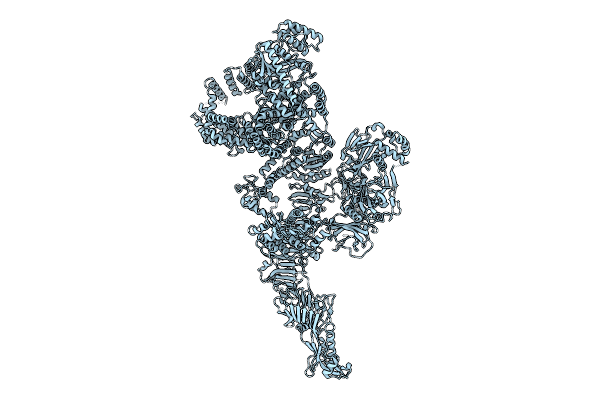
Organism: Escherichia coli o127:h6
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TOXIN Ligands: MN |
 |
Organism: Escherichia coli o127:h6
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TOXIN |
 |
Organism: Escherichia coli o127:h6
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TOXIN |
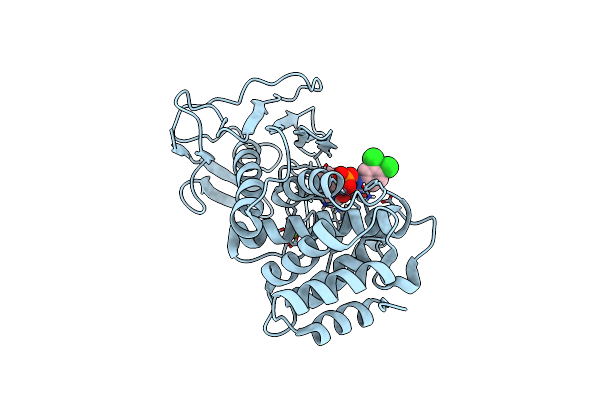 |
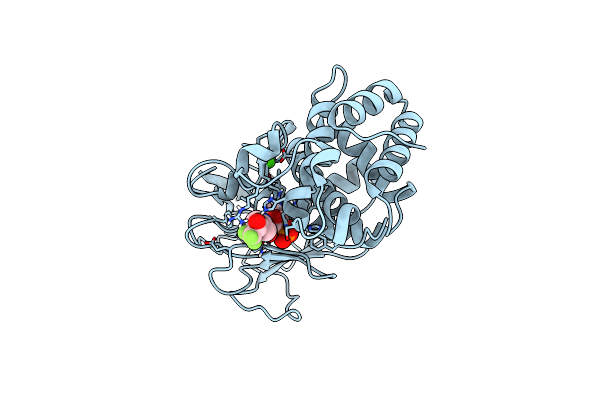
Pseudomonas Aeruginosa Elastase In Complex With A Phosphonate Based Inhibitor (S-Configured)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: CA, ZN, A1IJJ |
 |
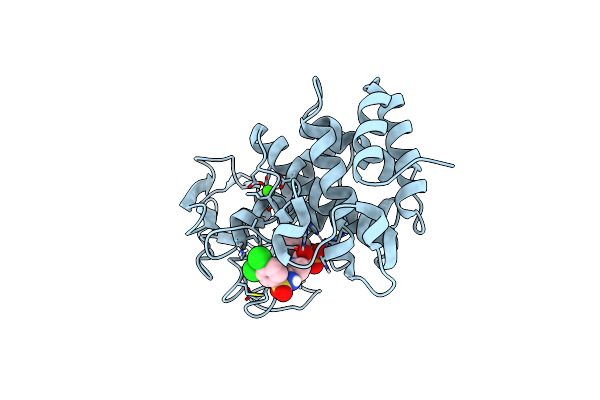
Pseudomonas Aeruginosa Elastase In Complex With A Phosphonate Based Inhibitor (R-Configured)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: ZN, CA, A1IFN |
 |
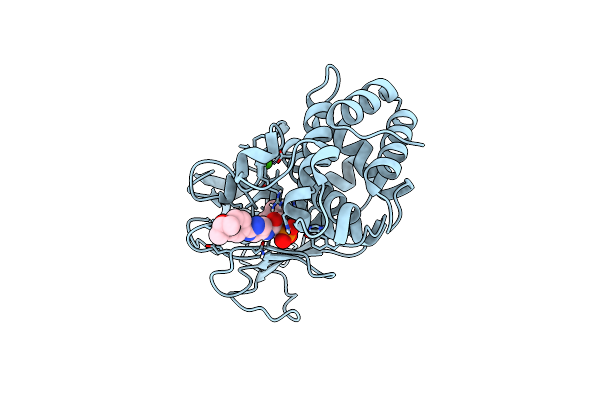
Pseudomonas Aeruginosa Elastase In Complex With A Phosphonate Based Inhibitor (R-Configured)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: A1IFZ, CA, ZN |
 |
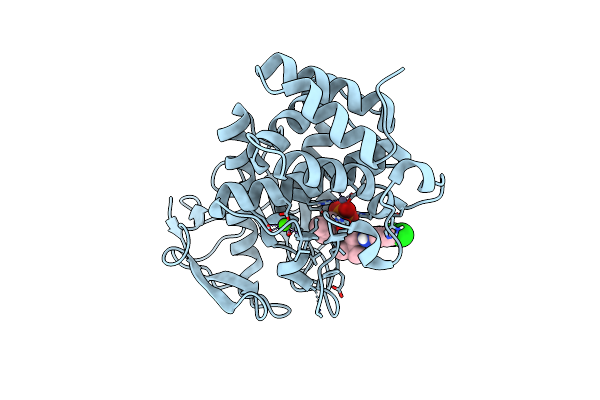
Pseudomonas Aeruginosa Elastase In Complex With A Phosphonate Based Inhibitor (R-Configured)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: CA, ZN, A1IFY |
 |
Pseudomonas Aeruginosa Elastase In Complex With A Phosphonate Based Inhibitor (R-Configured)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE Ligands: ZN, CA, A1IEX |
 |
Pseudomonas Aeruginosa Elastase In Complex With A Phosphonate Based Inhibitor (R-Configured)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE Ligands: CA, A1IEU, ZN |
 |
Organism: Escherichia coli o127:h6 str. e2348/69
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: TOXIN |
 |
Organism: Escherichia coli o127:h6 str. e2348/69
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: TOXIN |
 |
Organism: Treponema denticola
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: E64, EDO |

