Search Count: 2,800
All
Selected
 |
Cryo-Em Structure Of The Glucose-Specific Pts Transporter Iic From V. Cholerae In The Inward-Facing Conformation
Organism: Vibrio cholerae m66-2
Method: ELECTRON MICROSCOPY Resolution:3.68 Å Release Date: 2026-01-14 Classification: TRANSPORT PROTEIN |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: GOL, SO4 |
 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment And 5-Mer Rna
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
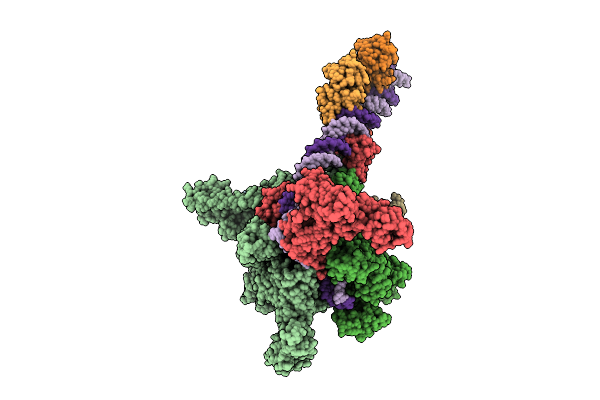
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr Transcription Factor And Ompu Promoter Dna
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Tcpp Transcription Factor And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
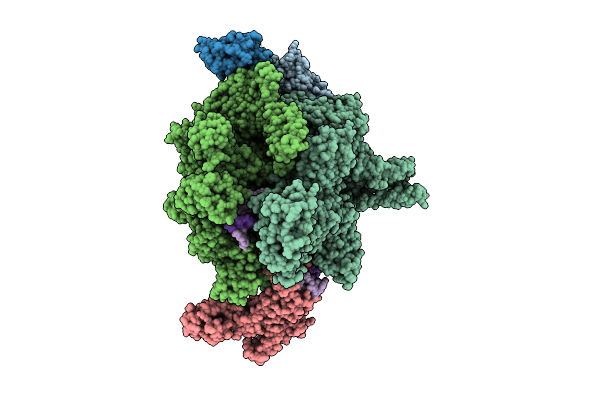 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr And Tcpp Transcription Factors And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Resolution:3.13 Å Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Resolution:2.93 Å Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN/DNA |
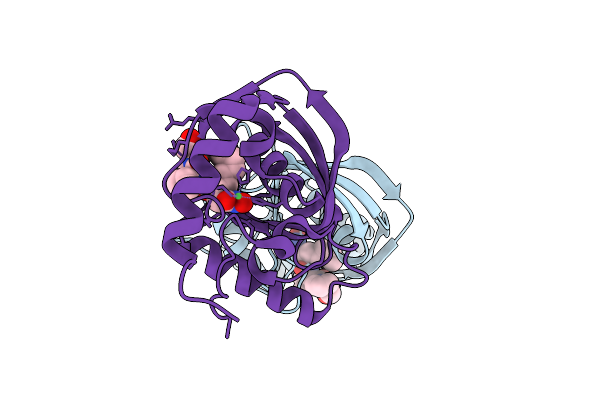 |
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NI, BB2 |
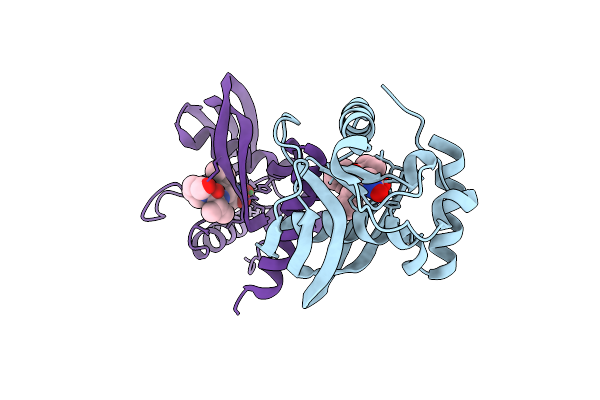 |
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: BB2, NI |
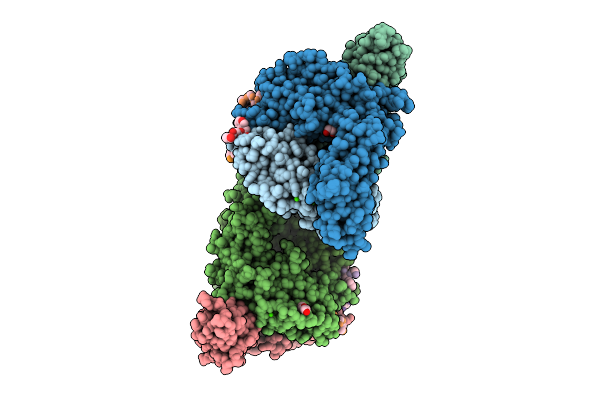 |
Vibrio Cholerae Protein Frha Peptid-Binding Domain And Adjacent Split Domain (S1127-F1439) In Complex With Peptide Agwtd X-Ray Crystallography Structure
Organism: Vibrio cholerae o395, Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: CA, PEG, EDO |
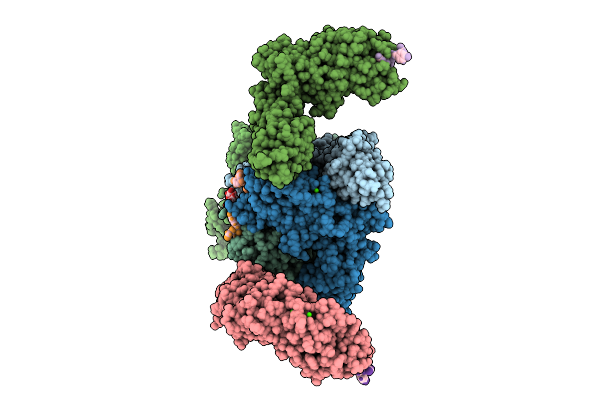 |
Vibrio Cholerae Protein Frha Peptid-Binding Domain And Adjacent Split Domain (S1127-F1439) In Complex With Peptide Agytd X-Ray Crystallography Structure
Organism: Vibrio cholerae o395, Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: CA |
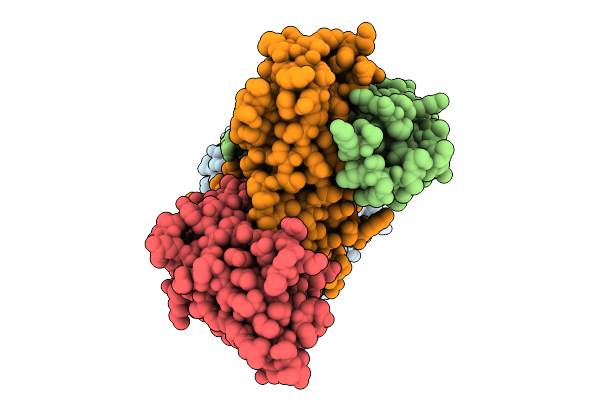 |
Organism: Vibrio cholerae, Synthetic construct, Lama glama
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: DM2 |
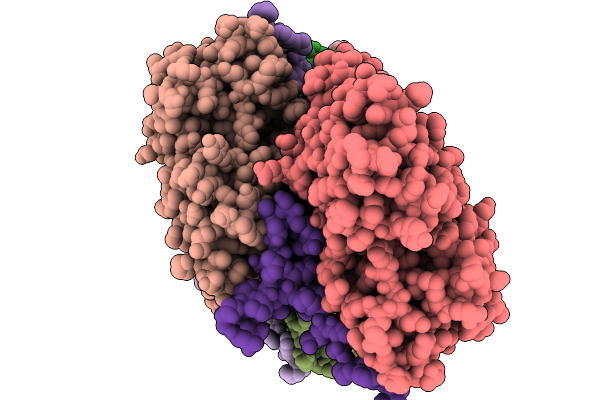 |
Cryo-Em Structure Of Vibrio Effector Vopv Fragment Bound To Skeletal Alpha F-Actin
Organism: Oryctolagus cuniculus, Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: STRUCTURAL PROTEIN Ligands: MG, ANP |
 |
Cryo-Em Structure Of The Type Ii Secretion System Protein From Vibrio Cholerae
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTEIN TRANSPORT |
 |
Vibrio Cholerae Cytolysin Cradle Loop Mutant-Y194A Half Barrel Pre-Pore Model
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: TOXIN |

