Search Count: 17
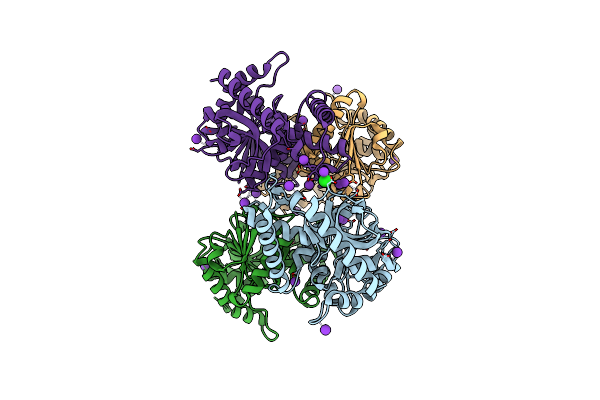 |
Crystal Structure Of Malate Dehydrogenase From Haloarcula Marismortui With Potassium And Chloride Ions
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-16 Classification: OXIDOREDUCTASE Ligands: CL, K |
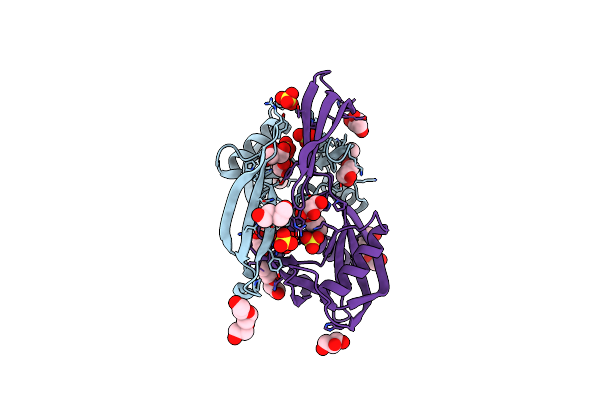 |
Crystal Structure Of The Bdellovibrio Bacteriovorus Nucleoside Diphosphate Sugar Hydrolase
Organism: Bdellovibrio bacteriovorus
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2016-01-20 Classification: HYDROLASE Ligands: SO4, GOL, PEG, PG0, PGE |
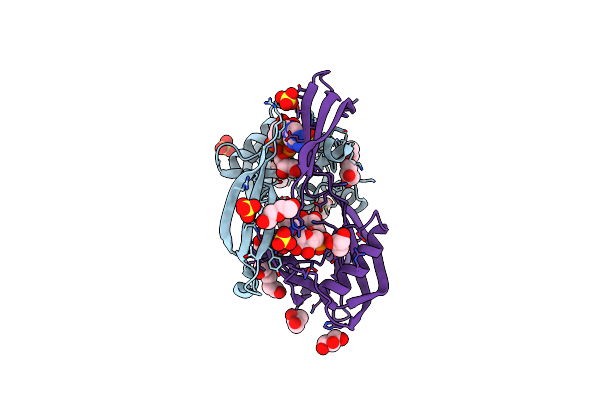 |
Crystal Structure Of The Bdellovibrio Bacteriovorus Nucleoside Diphosphate Sugar Hydrolase In Complex With Adp-Ribose
Organism: Bdellovibrio bacteriovorus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2016-01-20 Classification: HYDROLASE Ligands: APR, SO4, GOL, PG0, PEG, PGE |
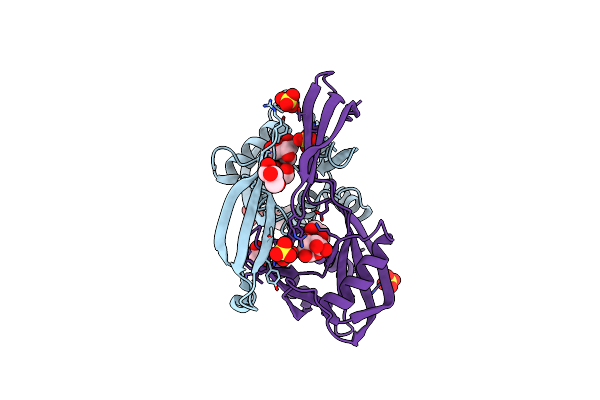 |
Crystal Structure Of The Bdellovibrio Bacteriovorus Nucleoside Diphosphate Sugar Hydrolase
Organism: Bdellovibrio bacteriovorus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-01-20 Classification: HYDROLASE Ligands: GLC, SO4, GOL, PEG |
 |
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-06-08 Classification: BIOSYNTHETIC PROTEIN Ligands: 3O3, BR |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-02-02 Classification: HYDROLASE Ligands: UA3 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2005-02-02 Classification: HYDROLASE Ligands: UM3 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2005-02-02 Classification: HYDROLASE Ligands: UMF |
 |
Crystal Structure At 1.8 Angstroms Of Seleno Methionyled Crh, The Bacillus Subtilis Catabolite Repression Containing Protein Crh Reveals An Unexpected Swapping Domain As An Untertwinned Dimer
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-10-07 Classification: TRANSPORT PROTEIN Ligands: SO4, GOL |
 |
Crystal Structure At 1.8 Angstroms Of The Bacillus Subtilis Catabolite Repression Histidine Containing Protein (Crh)
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-10-07 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-07-22 Classification: CELLULASE DEGRADATION Ligands: CA |
 |
Organism: Erwinia chrysanthemi
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1999-03-26 Classification: HYDROLASE Ligands: CA |
 |
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1999-03-16 Classification: HYDROLASE INHIBITION Ligands: CA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1999-01-13 Classification: COMPLEX (PROTEIN TRANSPORT/PEPTIDE) |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1999-01-06 Classification: PROTEIN TRANSPORT Ligands: CO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1998-12-23 Classification: GENE REGULATION |
 |
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-06-03 Classification: GLYCOSYL HYDROLASE Ligands: CA, ZN |

