Search Count: 173
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: PROTEIN TRANSPORT Ligands: MAN, NAG |
 |
Self Assembly Domain Of The Surface Layer Protein Of Viridibacillus Arvi (Aa 765-844)
Organism: Viridibacillus arvi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-08-28 Classification: STRUCTURAL PROTEIN Ligands: BR |
 |
Cell Wall Anchoring Domain Of The Surface Layer Protein Of Methanococcus Voltae (Aa 24-75; 484-576)
Organism: Methanococcus voltae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-08-28 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Alid In Open Conformation
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN Ligands: MG |
 |
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Alid In Closed Conformation In Complex With Peptide 1
Organism: Streptococcus pneumoniae, Prevotella
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Alic As A Domain-Swapped Dimer
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN |
 |
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Alib In Complex With An Unknown Peptide
Organism: Streptococcus pneumoniae, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN |
 |
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Alib In Complex With Peptide 2
Organism: Streptococcus pneumoniae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN |
 |
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Alib In Complex With Peptide 3
Organism: Streptococcus pneumoniae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN |
 |
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Alib In Complex With Peptide 4
Organism: Streptococcus pneumoniae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN |
 |
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Amia In Complex With Peptide 5
Organism: Streptococcus pneumoniae, Bacteria
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-05-22 Classification: PEPTIDE BINDING PROTEIN |
 |
Crystal Structure Of The Pneumococcal Substrate-Binding Protein Amia In Complex With An Unknown Peptide
Organism: Streptococcus pneumoniae, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-12-20 Classification: PEPTIDE BINDING PROTEIN |
 |
Heterodimer Formation Of Sensory Domains Of Vibrio Cholerae Regulators Toxr And Toxs
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-09-13 Classification: MEMBRANE PROTEIN |
 |
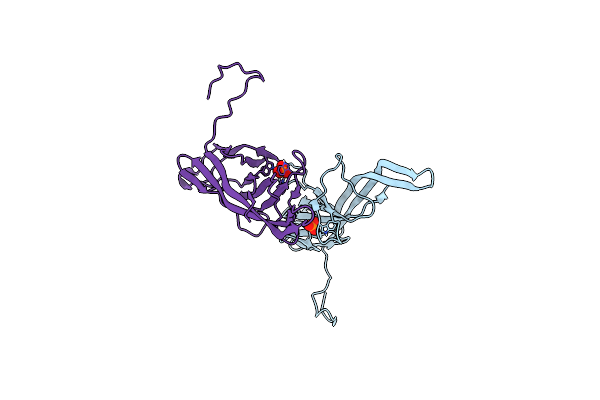
Crystal Structure Of Slpa Domain Ii (Aa 201-310), Domain That Is Involved In The Self-Assembly And Dimerization Of The S-Layer From Lactobacillus Acidophilus
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-09-06 Classification: STRUCTURAL PROTEIN Ligands: NCA |
 |
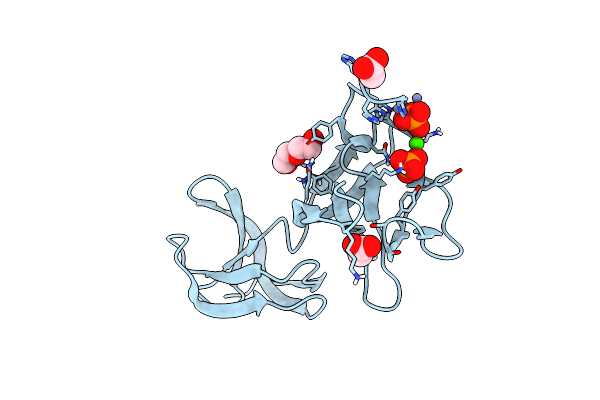
S-Layer Protein Slpa From Lactobacillus Amylovorus, Domain I (Aa 32-209), Important For Self-Assembly
Organism: Lactobacillus amylovorus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-09-06 Classification: STRUCTURAL PROTEIN Ligands: PO4 |
 |
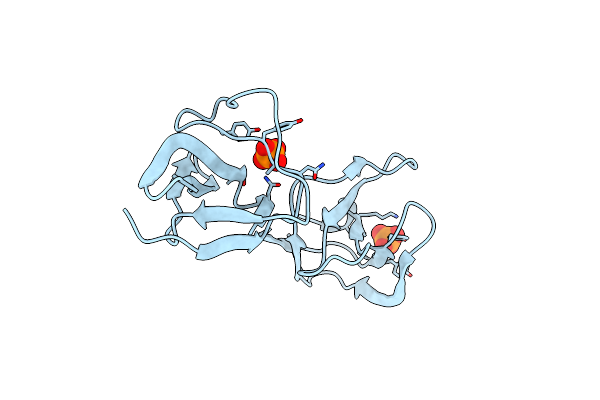
Crystal Structure Of S-Layer Protein Slpx From Lactobacillus Acidophilus, Domain Iii (Aa 363-499)
Organism: Lactobacillus acidophilus atcc 4796
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-08-23 Classification: CELL ADHESION Ligands: ZN, PO4, CA, ACT, PEG |
 |
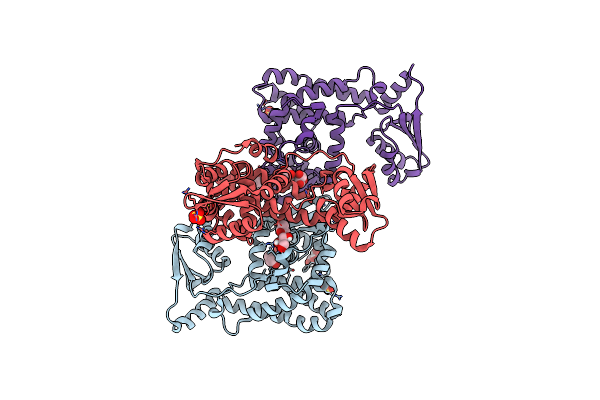
Crystal Structure Of The Teichoic Acid Binding Domain Of Slpa, S-Layer Protein From Lactobacillus Acidophilus (Aa. 314-444)
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-08-16 Classification: CELL ADHESION Ligands: PO4 |
 |
Organism: Escherichia coli w
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-07-05 Classification: TRANSCRIPTION Ligands: GOL, SO4 |
 |
Crystal Structure Of S-Layer Protein Slpa From Lactobacillus Acidophilus, Domain I, Co-Crystallization With Hgcl2, Mutation Ser146Cys, (Aa 32-198)
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-12-28 Classification: STRUCTURAL PROTEIN Ligands: HG, CL |
 |
Crystal Structure Of S-Layer Protein Slpa From Lactobacillus Acidophilus, Domain I (Aa 32-198)
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-12-28 Classification: STRUCTURAL PROTEIN |

