Search Count: 20
 |
Crystal Structure Of Aminoglycoside Tc007 In Complex With 70S Ribosome From Thermus Thermophilus, Three Trnas And Mrna (Soaking)
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2018-05-02 Classification: RIBOSOME Ligands: 8UZ, MG, ZN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2017-12-13 Classification: RIBOSOME Ligands: MG, GET, ZN |
 |
Crystal Structure Of Aminoglycoside Tc007 Co-Crystallized With 70S Ribosome From Thermus Thermophilus, Three Trnas And Mrna
Organism: Escherichia coli, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2017-12-13 Classification: RIBOSOME Ligands: 8UZ, MG, ZN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain rm11-1a)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-12-13 Classification: RIBOSOME Ligands: PAR, MG, ZN |
 |
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2017-12-13 Classification: RIBOSOME Ligands: MG, 8UZ, ZN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2017-12-13 Classification: RIBOSOME Ligands: MG, LLL, ZN |
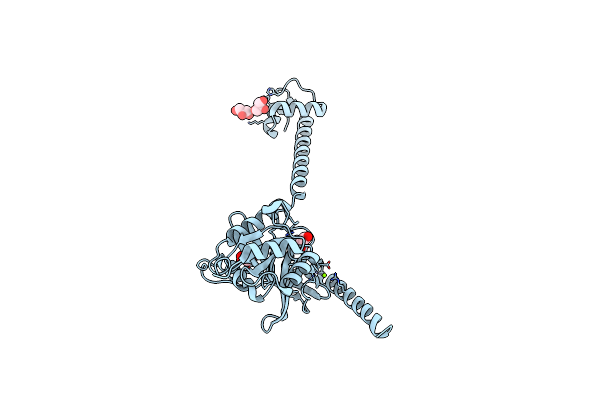 |
Bacterial Translation Initiation Factor If2 (1-363), Apo Form, Double Mutant K86L H130A
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2013-09-18 Classification: TRANSLATION Ligands: MG, GOL, IOD |
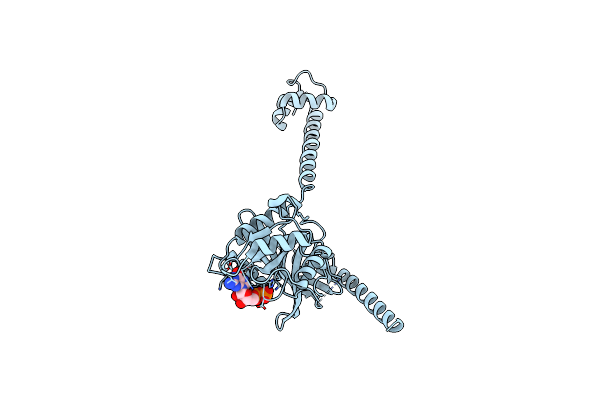 |
Bacterial Translation Initiation Factor If2 (1-363), Complex With Gdp At Ph8.0
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-09-18 Classification: TRANSLATION Ligands: GDP |
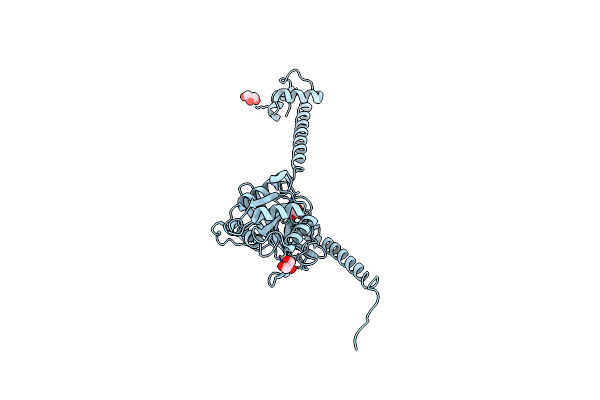 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-05-29 Classification: TRANSLATION Ligands: GOL |
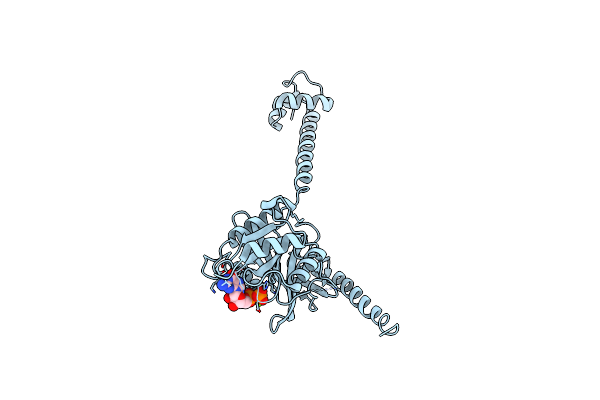 |
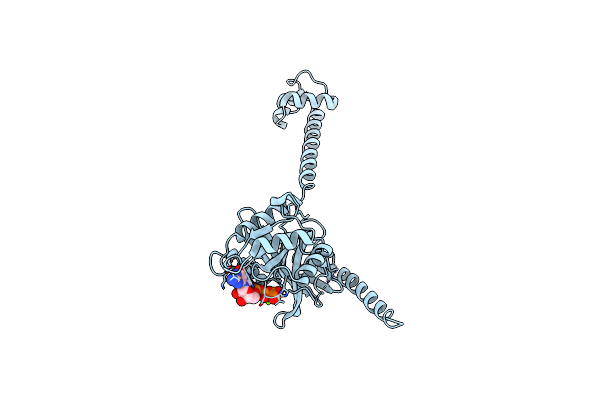
Bacterial Translation Initiation Factor If2 (1-363), Complex With Gdp At Ph6.5
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-05-29 Classification: TRANSLATION Ligands: GDP, MG |
 |
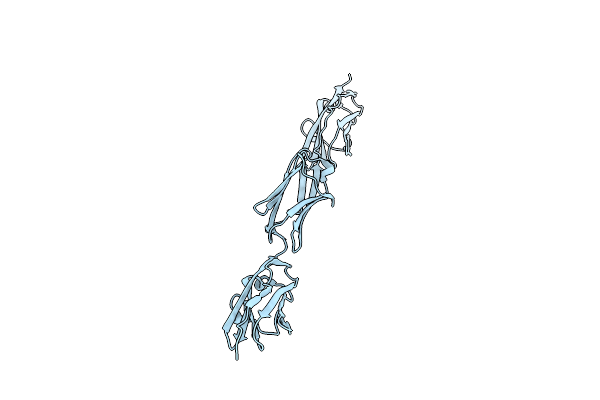
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-05-29 Classification: TRANSLATION Ligands: GTP, MG |
 |
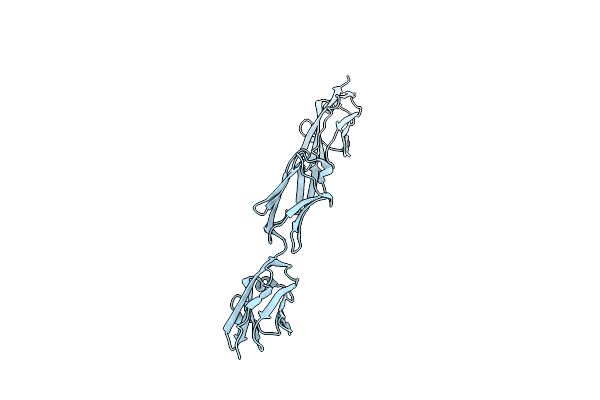
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-01-22 Classification: STRUCTURAL PROTEIN |
 |
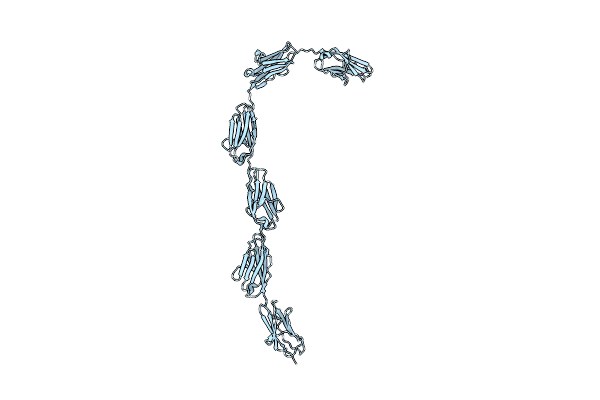
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-01-22 Classification: STRUCTURAL PROTEIN |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2008-01-22 Classification: STRUCTURAL PROTEIN |
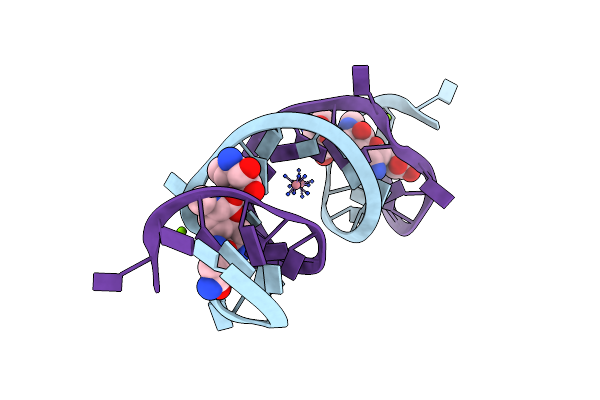 |
Crystal Structure Of The Homo Sapiens Cytoplasmic Ribosomal Decoding Site Complexed With Apramycin
Method: X-RAY DIFFRACTION
Resolution:2.80 Å Release Date: 2006-06-20 Classification: RNA Ligands: AM2, MG, NCO |
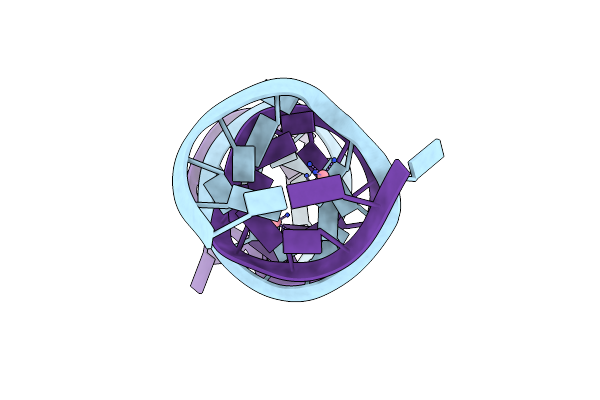 |
Crystal Structure Of The Homo Sapiens Cytoplasmic Ribosomal Decoding A Site
Method: X-RAY DIFFRACTION
Resolution:2.30 Å Release Date: 2006-02-14 Classification: RNA Ligands: MG, NCO |
 |
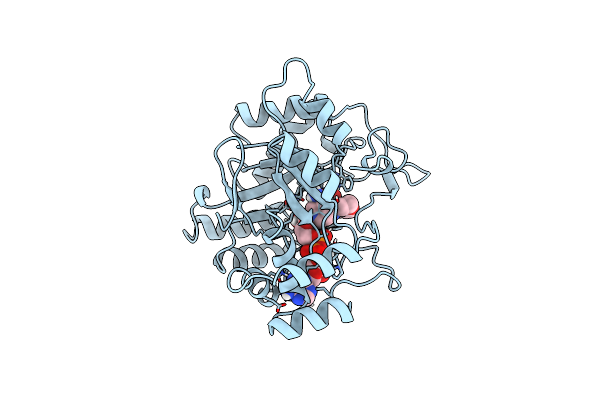
Dimerization Of E. Coli Dna Gyrase B Provides A Structural Mechanism For Activating The Atpase Catalytic Center
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2000-03-31 Classification: ISOMERASE Ligands: SO4, ANP, GOL |
 |
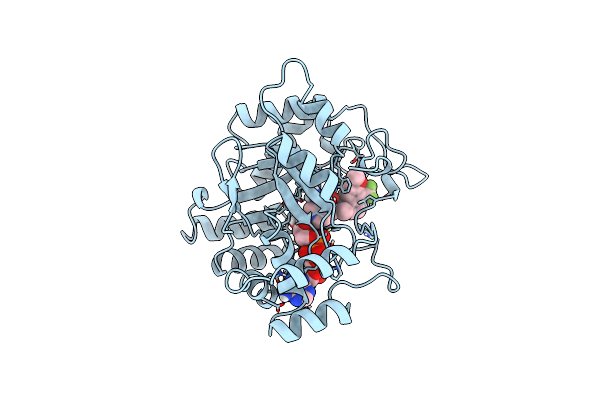
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1998-04-15 Classification: OXIDOREDUCTASE Ligands: NAP, SBI |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1998-04-15 Classification: OXIDOREDUCTASE Ligands: NAP, TOL |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-04-15 Classification: OXIDOREDUCTASE Ligands: NAP |

