Search Count: 209
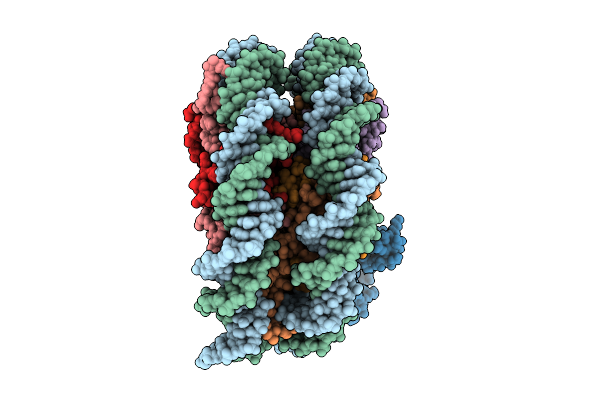 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION |
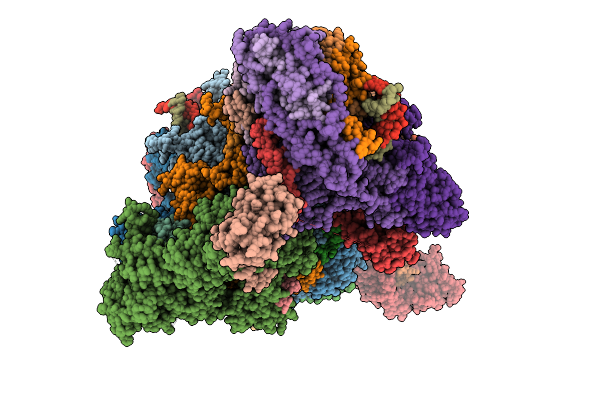 |
State 1 Map3 Rna Pol Ii Activated Elongation Complex With Setd2 And Upstream Hexasome
Organism: Homo sapiens, Synthetic construct, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
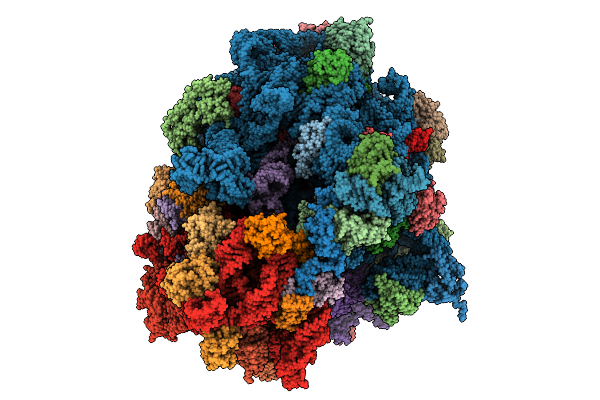
Structure Of The 70S-Ef-G(P610L)-Gdp-Pi Ribosome Complex With Trnas In Hybrid State 2 (H2-Ef-G(P610L)-Gdp-Pi)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: MG, ZN, NA, AM2, GDP, PO4 |
 |
Structure Of The 70S-Ef-G(P610L)-Gdp-Pi Ribosome Complex With Trnas In Hybrid State 1 (H1-Ef-G(P610L)-Gdp-Pi)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: RIBOSOME Ligands: MG, ZN, NA, AM2, PO4, GDP |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSCRIPTION |
 |
State 3 Map 3 Rna Pol Ii Activated Elongation Complex With Setd2 Bound To Distal Upstream H3
Organism: Homo sapiens, Synthetic construct, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Organism: Homo sapiens, Synthetic construct, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSCRIPTION Ligands: ZN, MG |
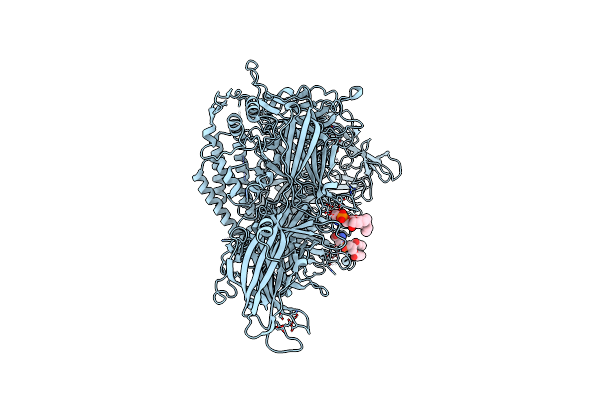 |
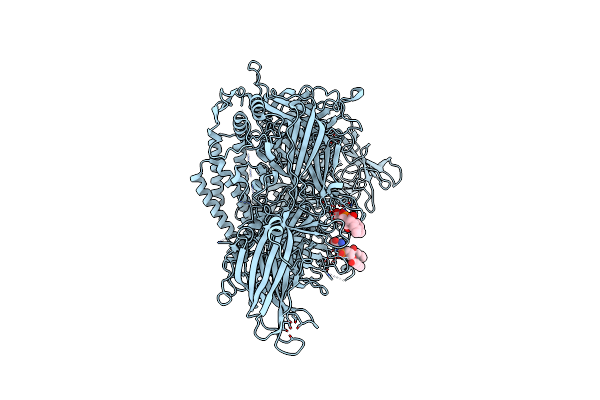
Cryo-Em Structure Of Lipid-Bound Human Myoferlin (25 Mol% Dops/5 Mol% Pi(4,5)P2 Nanodisc)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: PSF, CA |
 |
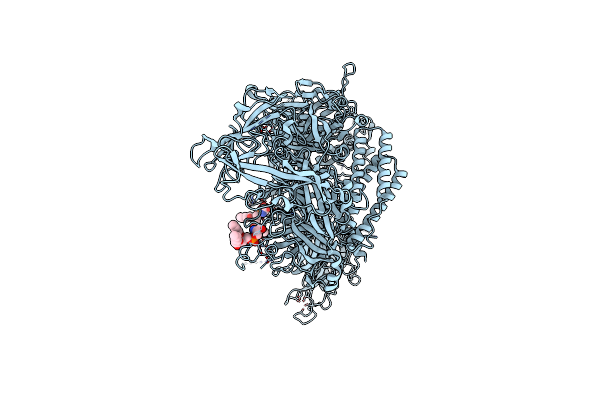
Human Myoferlin (1-1997) In Complex With An Msp2N2 Lipid Nanodisc (15 Mol% Dops, 5 Mol% Cholesterol)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: PSF, CA |
 |
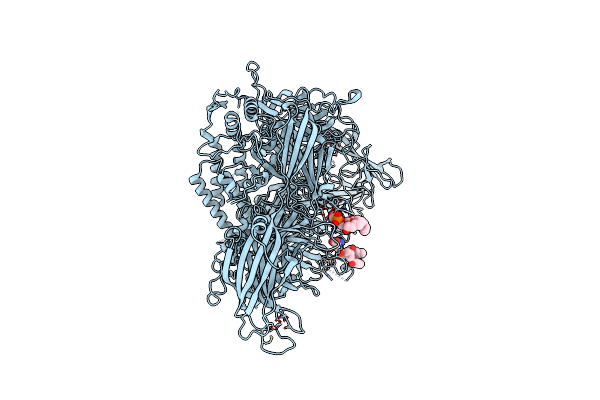
Human Myoferlin (1-1997) In Complex With An Msp2N2 Lipid Nanodisc (15 Mol% Dops, 2 Mol% Pi(4,5)P2)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: PSF, CA |
 |
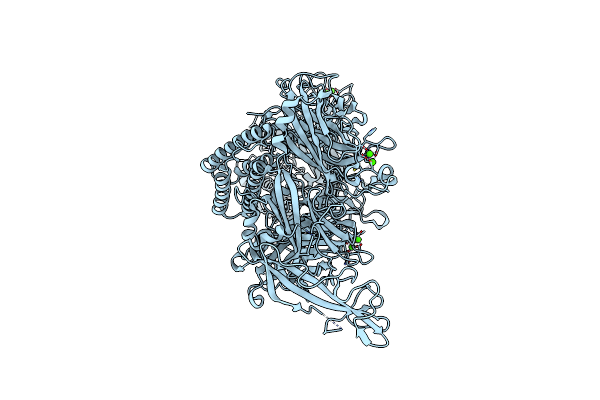
Human Myoferlin (1-1997) In Complex With An Msp2N2 Lipid Nanodisc (25 Mol% Dops, 5 Mol% Pi(4,5)P2 And 5 Mol% Cholesterol)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: PSF, CA |
 |
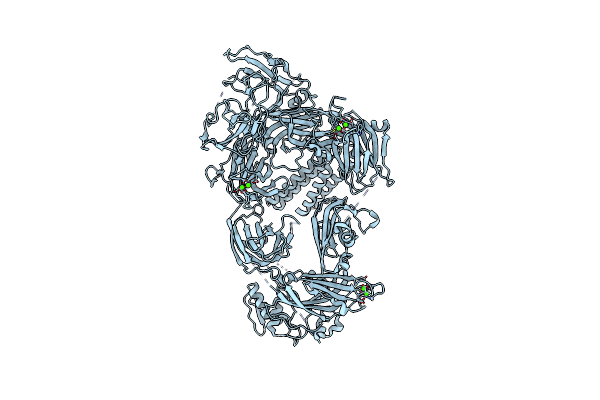
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
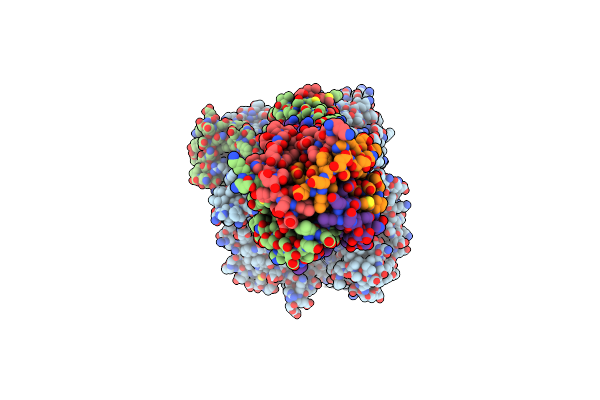
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
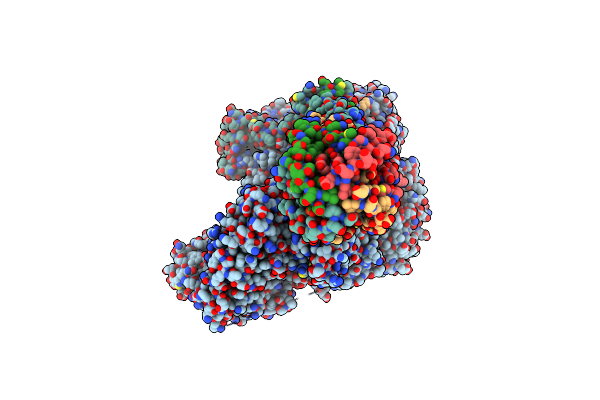
Organism: Henipavirus nipahense
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: ZN |
 |
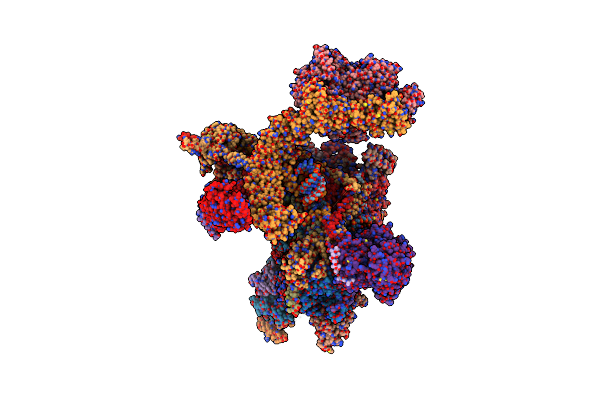
Structure Of Replicating Nipah Virus Rna Polymerase Complex - Rna-Bound State
Organism: Henipavirus nipahense
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: GNP, ZN, MG |
 |
Organism: Schizosaccharomyces pombe
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: SPLICING Ligands: MG, K, IHP, GTP, ZN |
 |
Organism: Schizosaccharomyces pombe
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: SPLICING Ligands: IHP, GTP, MG, ZN, K |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2024-11-20 Classification: TRANSFERASE |

