Search Count: 65
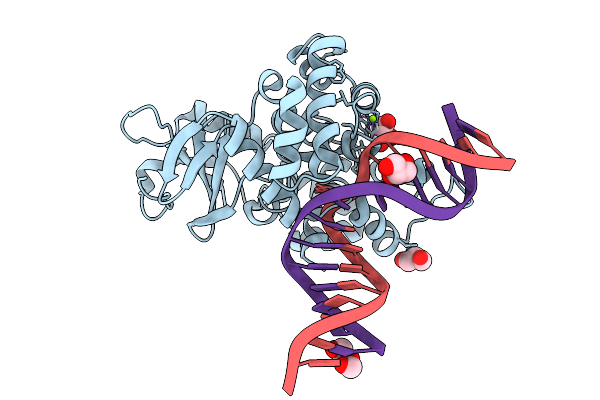 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Substrate 8-Oxoguanine Dna At Ph 8.0 Under 277 K
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: PEG, GOL, MG, NA |
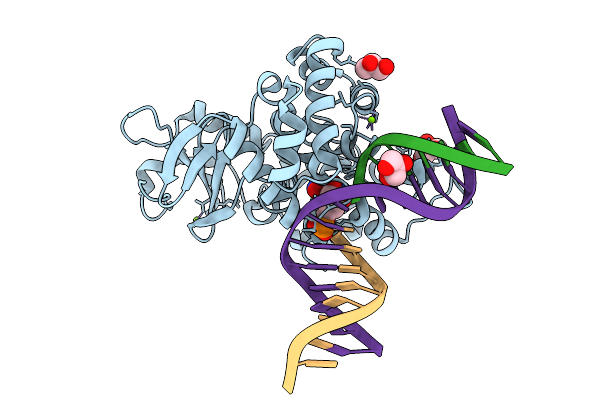 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 24 Hourss
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
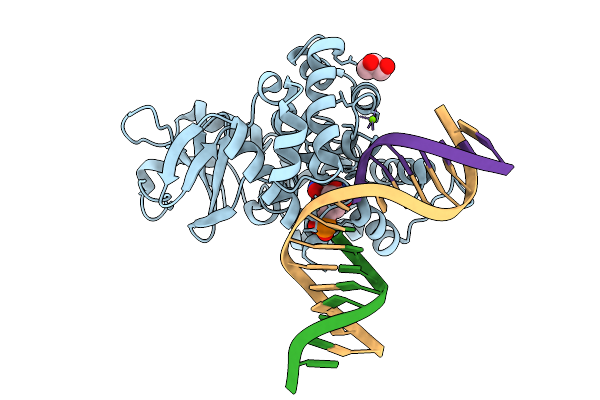 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 2.5 Hours
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
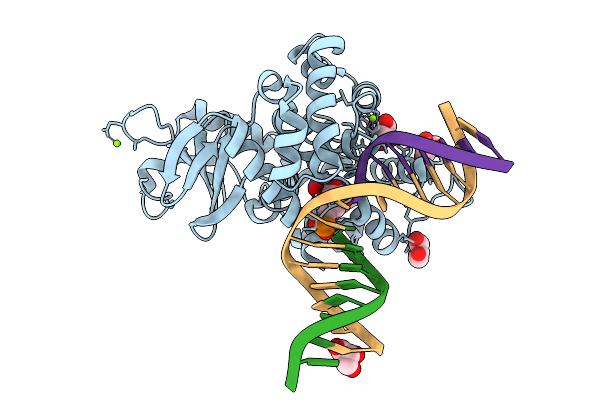 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 298 K For 3 Weeks
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, GOL, MG, NA |
 |
Organism: Microchaete diplosiphon
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-05-15 Classification: SIGNALING PROTEIN Ligands: CYC |
 |
Neutron Structure Of Bacillus Thermoproteolyticus Ferredoxin At Room Temperature
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.45 Å, 1.60 Å Release Date: 2024-02-07 Classification: ELECTRON TRANSPORT Ligands: SF4 |
 |
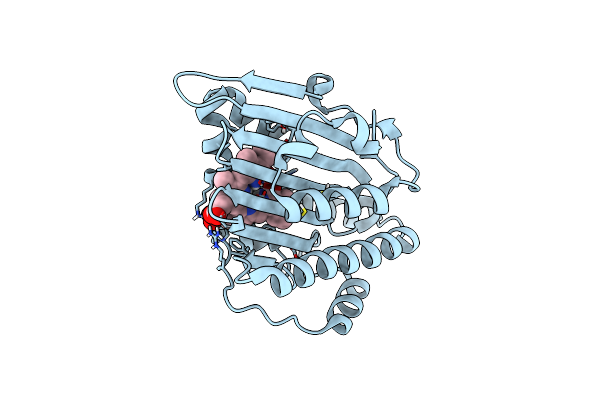
Crystal Structure Of The Hen Egg Lysozyme-2-Oxidobenzylidene-Threoninato-Copper (Ii) Complex
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:0.92 Å Release Date: 2023-05-10 Classification: HYDROLASE Ligands: EDO, CL, JI6, NA |
 |
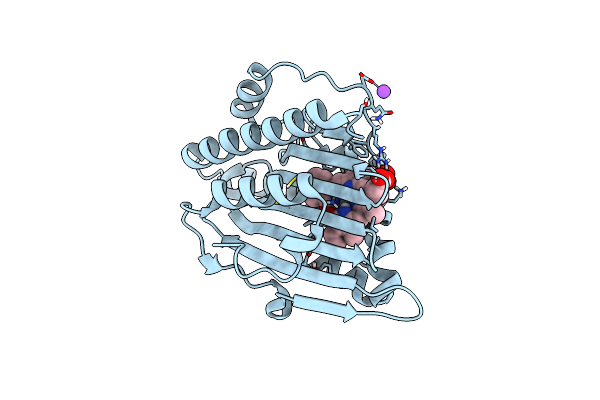
Neutron Structure Of Pcya I86D Mutant Complexed With Biliverdin At Room Temperature
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.90 Å, 2.0 Å Release Date: 2023-01-25 Classification: OXIDOREDUCTASE Ligands: BLR |
 |
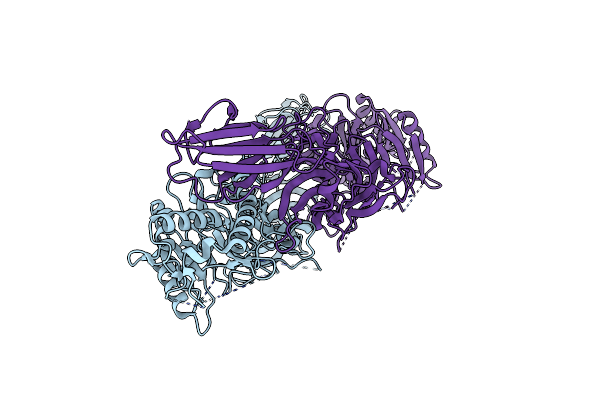
Neutron Structure Of Pcya D105N Mutant Complexed With Biliverdin At Room Temperature
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.38 Å, 2.10 Å Release Date: 2023-01-25 Classification: OXIDOREDUCTASE Ligands: BLR, NA |
 |
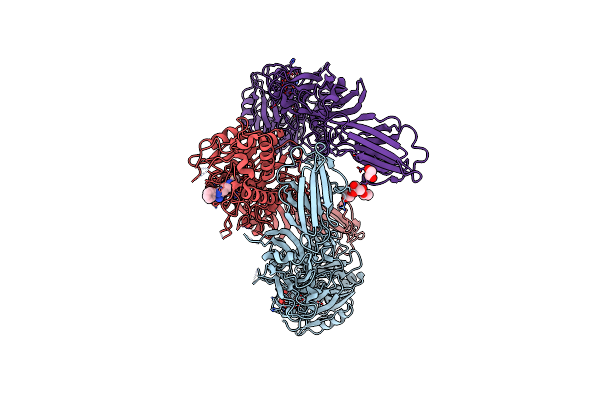
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2021-06-02 Classification: HYDROLASE |
 |
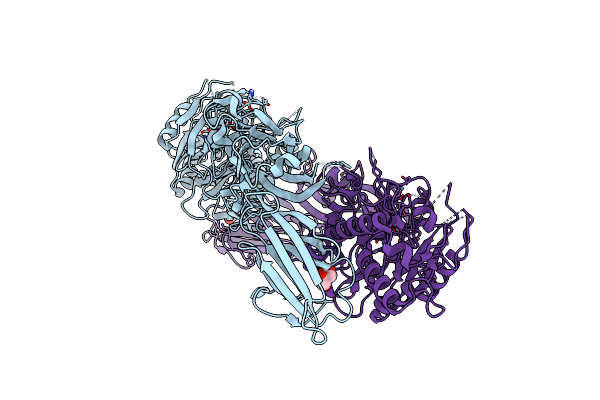
Structure Of The Peptidylarginine Deiminase Type Iii (Pad3) In Complex With Cl-Amidine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: BFB, CA, CL, EDO, GOL |
 |
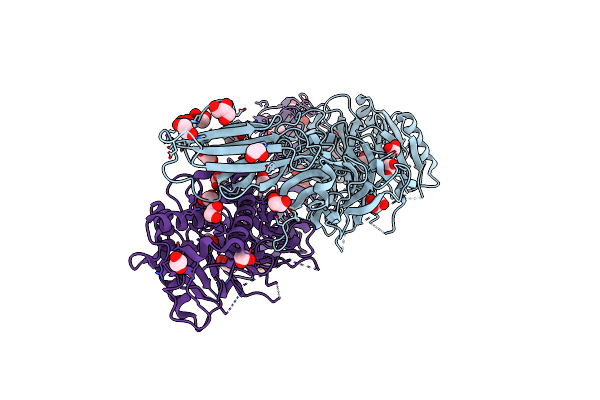
Structure Of The Ca2+-Bound C646A Mutant Of Peptidylarginine Deiminase Type Iii (Pad3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: CA, GOL, CL |
 |
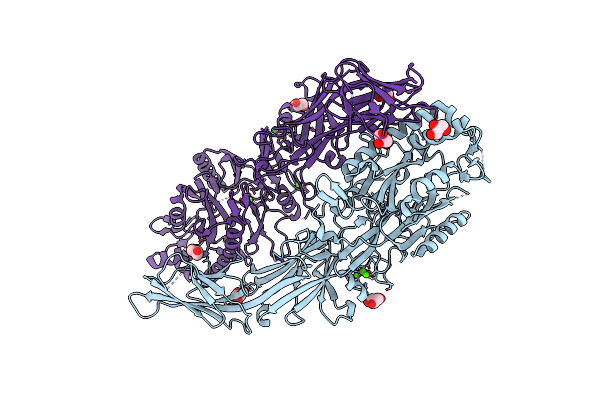
Structure Of The C646A Mutant Of Peptidylarginine Deiminase Type Iii (Pad3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: EDO, GOL |
 |
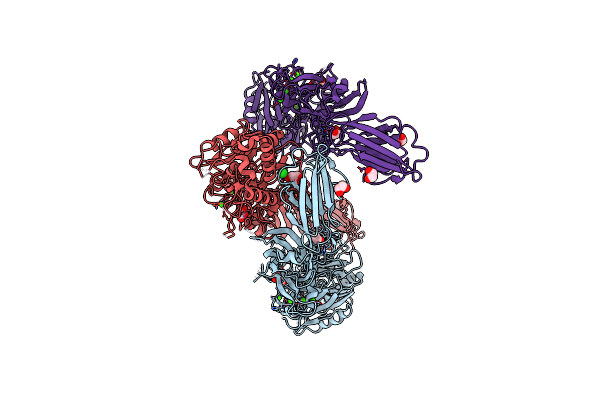
Structure Of The Inactive Form Of Wild-Type Peptidylarginine Deiminase Type Iii (Pad3) Crystallized Under The Condition With High Concentrations Of Ca2+
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: CA, GOL, CL |
 |
Structure Of The Ca2+-Bound Wild-Type Peptidylarginine Deiminase Type Iii (Pad3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: CA, CL, EDO, GOL |
 |
Organism: Microchaete diplosiphon
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-05-05 Classification: SIGNALING PROTEIN Ligands: CYC, MG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-04-07 Classification: CYTOKINE Ligands: GOL, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.47 Å Release Date: 2020-03-18 Classification: TRANSPORT PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-07-18 Classification: PROTEIN TRANSPORT Ligands: NAG, NA, ACT |
 |
Crystal Structure Of I86D Mutant Of Phycocyanobilin:Ferredoxin Oxidoreductase In Complex With Biliverdin (Data 1)
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2017-03-15 Classification: OXIDOREDUCTASE Ligands: BLA |

