Search Count: 578
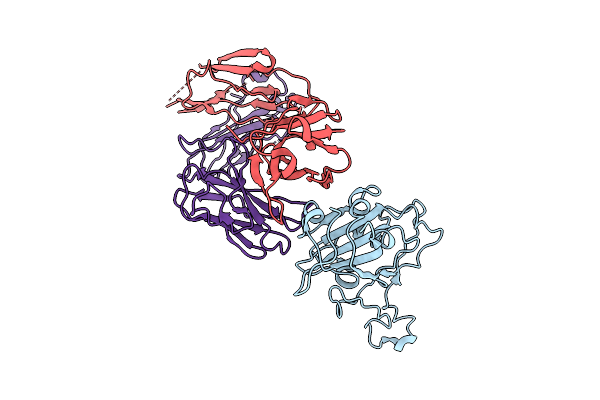 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
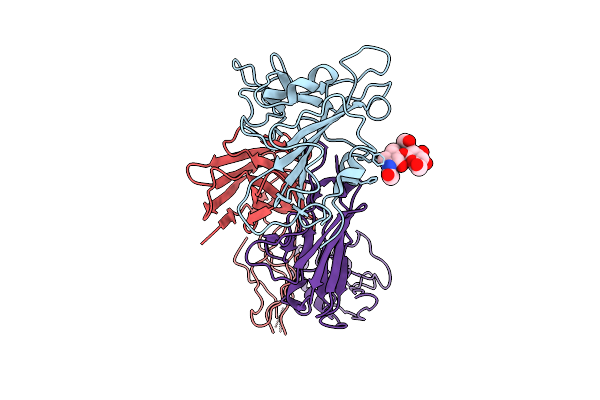 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-183
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
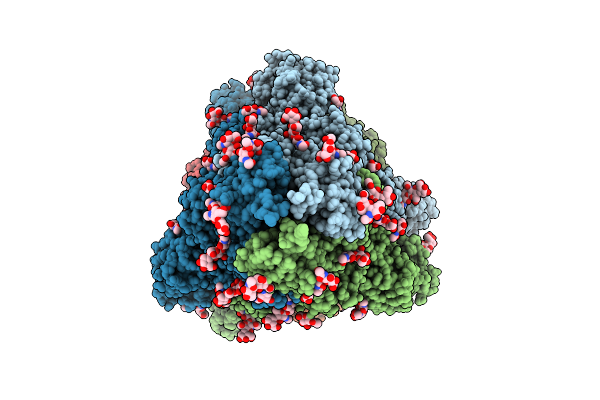 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
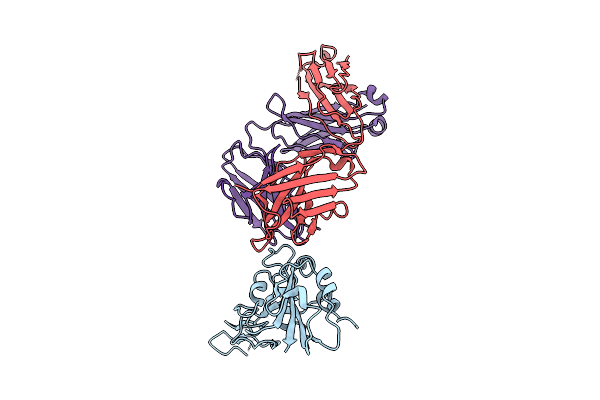 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
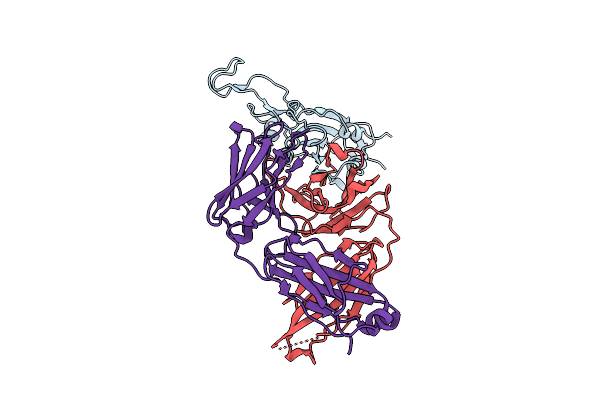 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
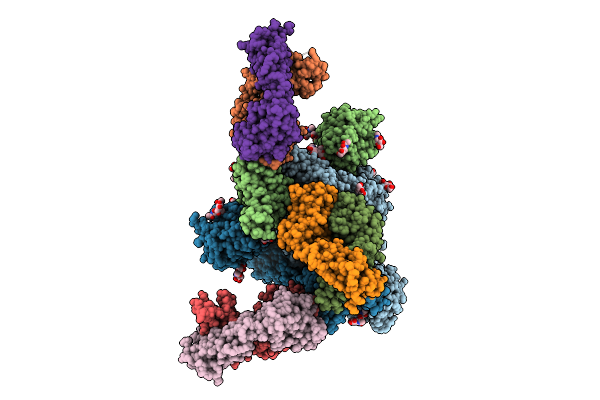 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
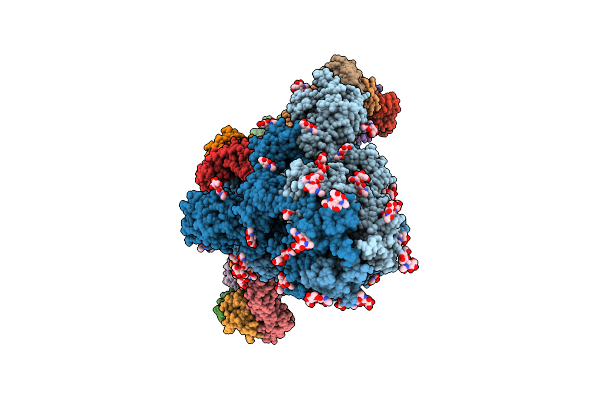 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
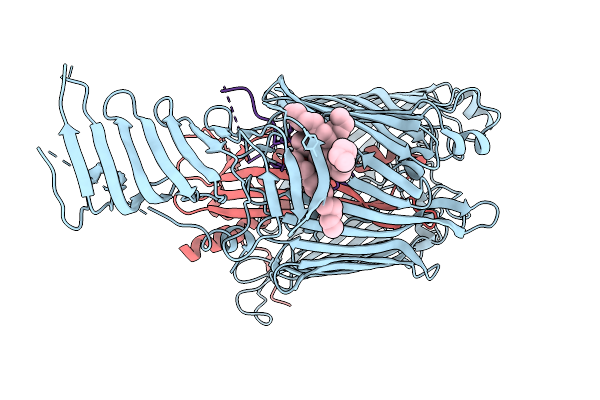 |
Cryo-Em Structure Of Lptdem Complex Containing Shigella Flexneri Lpte And Endogenous E. Coli Lptd And Lptm
Organism: Shigella flexneri, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: PLM, PXS |
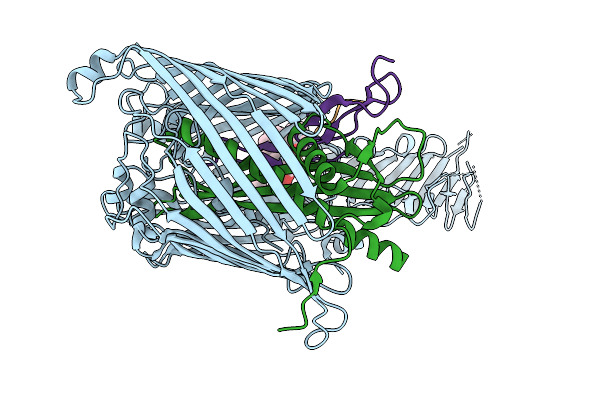 |
Cryo-Em Structure Of Shigella Flexneri Lptde In Complex With Rtp45 Superinfection Exclusion Protein From Rtp Bacteriophage And Endogenous Lptm
Organism: Shigella flexneri, Escherichia phage rtp, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: PLM, PXS |
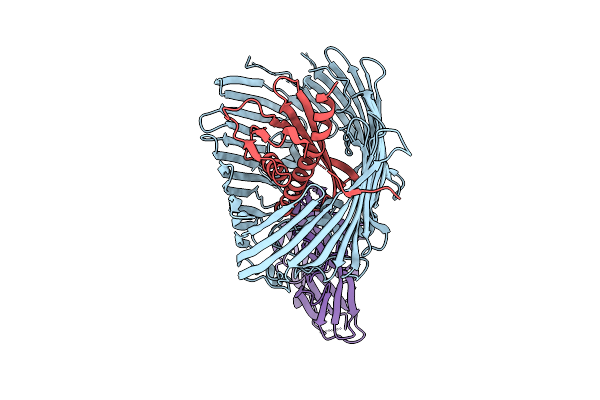 |
Cryo-Em Structure Of The Open State Of Shigella Flexneri Lptde Bound By The Rbp Of Oekolampad Phage
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
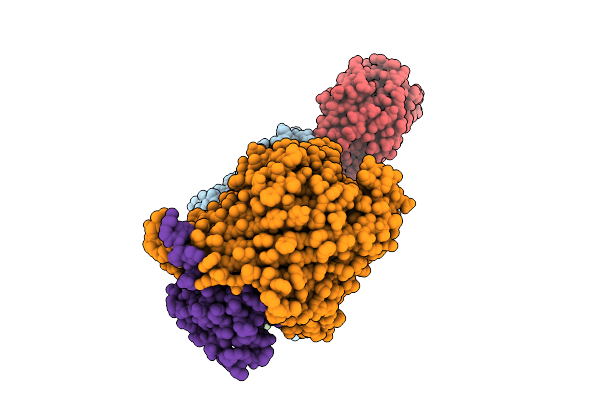 |
Cryo-Em Structure Of Shigella Flexneri Lptde Dimer: Closed-State Unbound And Open-State Bound By Oekolampad Phage Rbp
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
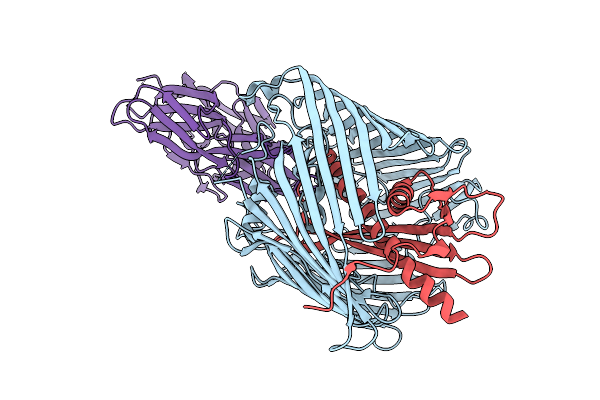 |
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By Phage Rbp Reveals N-Terminal Strand Insertion Into Lateral Gate
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
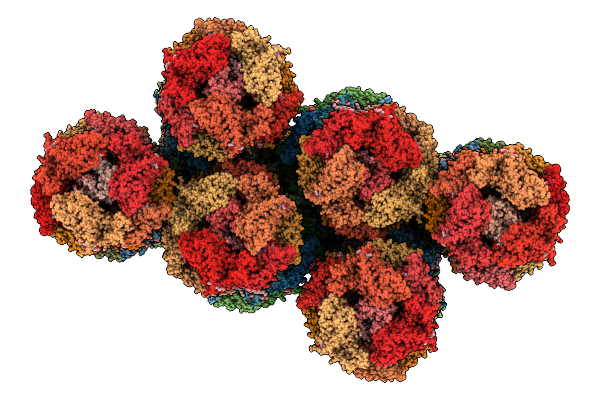 |
Organism: Gloeobacter violaceus pcc 7421
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
Cryoem Structure Of De Novo Antibody Fragment Scfv 6 With C. Difficile Toxin B (Tcdb)
Organism: Clostridioides difficile, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN |
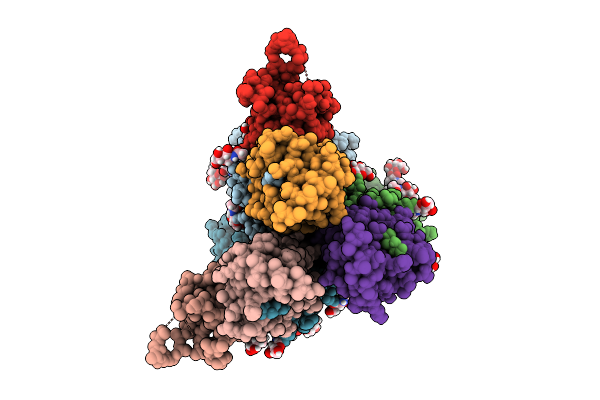 |
Cryoem Structure Of De Novo Vhh, Vhh_Flu_01, Bound To Influenza Ha, Strain A/Usa:Iowa/1943 H1N1.
Organism: Synthetic construct, Influenza a virus (strain a/usa:iowa/1943 h1n1)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN Ligands: NAG |
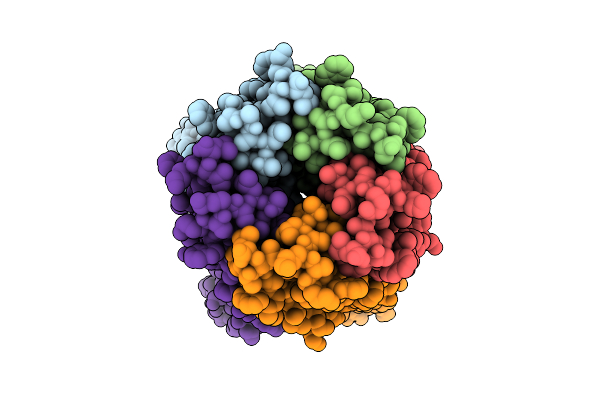 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DE NOVO PROTEIN Ligands: IOD |
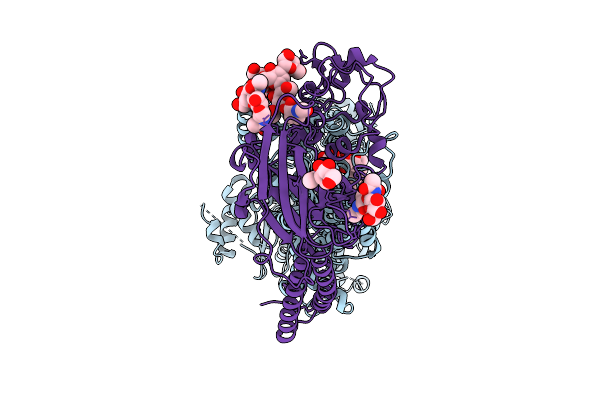 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
 |
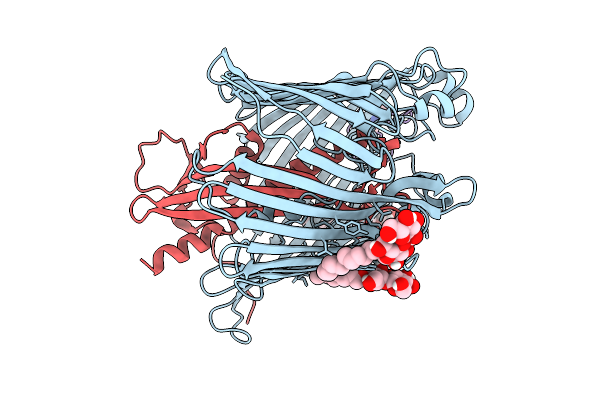
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By A Bicyclic Peptide Molecule (Compound 1)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, A1I1D |
 |
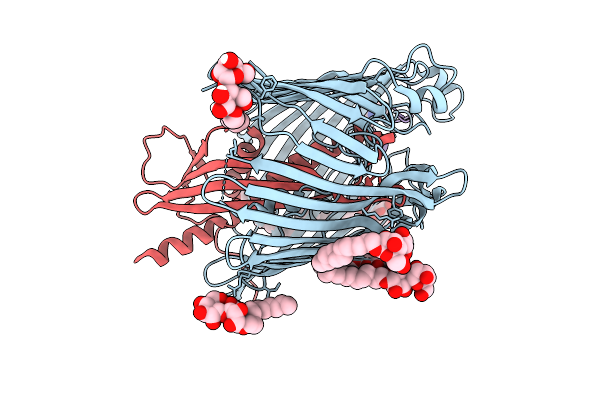
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By A Bicyclic Peptide Molecule (Compound 2)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, KZ0 |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By A Bicyclic Peptide Molecule (Compound 3)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, KZ0 |

