Search Count: 16
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-01-15 Classification: MOTOR PROTEIN Ligands: ADP, MG |
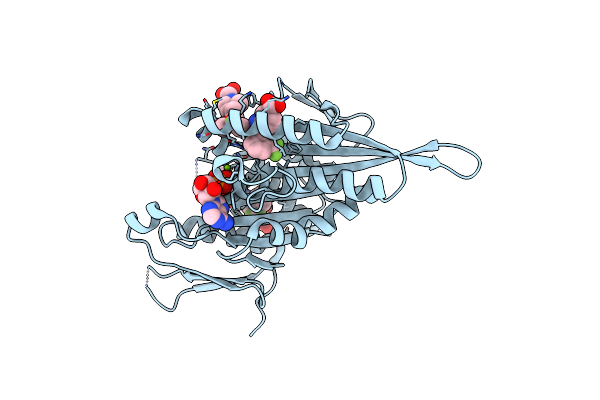 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2013-01-23 Classification: CELL CYCLE Ligands: ADP, MG, 4A2, EPE, PEG |
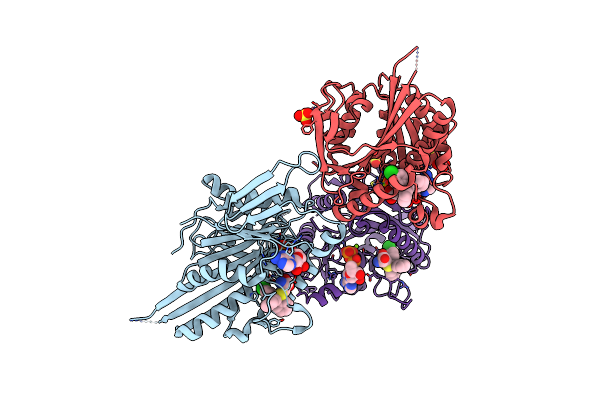 |
Crystal Structure Of Human Kinesin Eg5 In Complex With (R)-2-Amino-3-((S)-2-Methyl-1,1-Diphenylbutylthio)Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-03-30 Classification: CELL CYCLE Ligands: 2XA, MG, ADP, CL, SO4 |
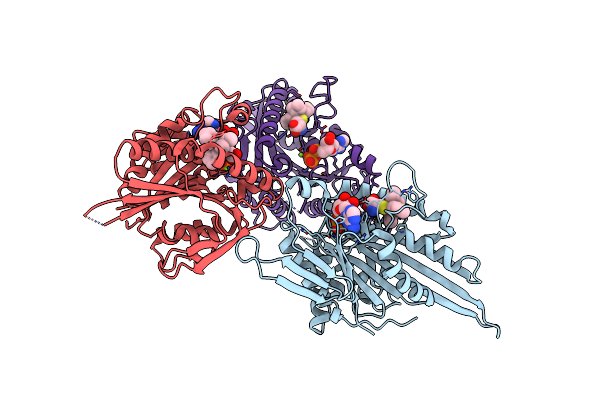 |
Crystal Structure Of Human Kinesin Eg5 In Complex With (R)-2-Amino-3-((4-Chlorophenyl)Diphenylmethylthio)Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-01-26 Classification: CELL CYCLE Ligands: ADP, X2O, MG |
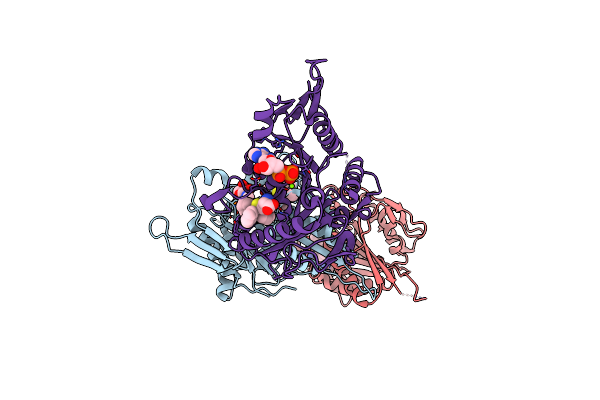 |
Intermediate And Final States Of Human Kinesin Eg5 In Complex With S-Trityl-L-Cysteine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-08-04 Classification: CELL CYCLE Ligands: ADP, MG, ZZD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-07-14 Classification: CELL CYCLE Ligands: ADP, MG, KZ9 |
 |
Crystal Structure Of Human Kinesin Eg5 In Complex With (S)-Dimethylenastron
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-07-14 Classification: CELL CYCLE Ligands: ADP, MG, EGB |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-07-14 Classification: CELL CYCLE Ligands: ADP, MG, X7E |
 |
Crystal Structure Of 2C-Methyl-D-Erythritol 2,4-Clycodiphosphate Synthase Complexed With Ligand
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-08-25 Classification: LYASE Ligands: ZN, F01, GPP |
 |
Crystal Structure Of 2C-Methyl-D-Erythritol 2,4-Clycodiphosphate Synthase Complexed With Ligand
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2009-08-25 Classification: LYASE Ligands: ZN, NA, CFV, GPP |
 |
Crystal Structure Of 2C-Methyl-D-Erythritol 2,4-Clycodiphosphate Synthase Complexed With Aracmp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-08-25 Classification: LYASE Ligands: ZN, CAR, SO4, GPP |
 |
Crystal Structure Of 2C-Methyl-D-Erythritol 2,4-Clycodiphosphate Synthase Complexed With Ligand
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-08-25 Classification: LYASE Ligands: ZN, CC7, MG, GPP |
 |
Crystal Structure Of 2C-Methyl-D-Erythritol 2,4-Clycodiphosphate Synthase Complexed With Ligand
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2009-08-25 Classification: LYASE Ligands: ZN, C6B, GPP, GOL |
 |
Crystal Structure Of Staphylococcus Aureus 1,4-Dihydroxy-2-Naphthoyl Coa Synthase (Menb) In Complex With Acetoacetyl Coa
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2007-11-13 Classification: LYASE Ligands: CAA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-04-24 Classification: TRANSFERASE Ligands: PEG, EDO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2007-04-24 Classification: TRANSFERASE Ligands: UD1 |

