Search Count: 32
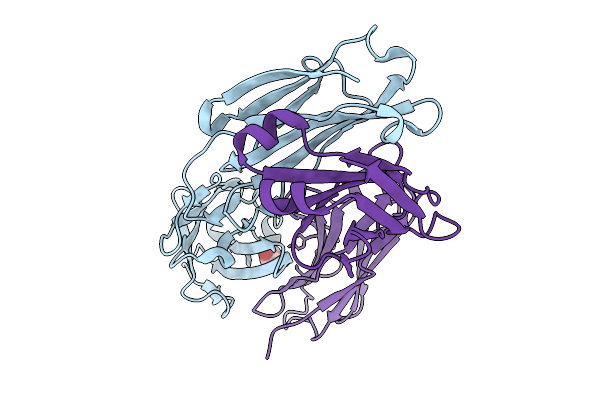 |
Crystal Structure Of Switchbody Based On Anti-Osteocalcin Antibody Ktm219 Fab
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: IMMUNE SYSTEM Ligands: GOL |
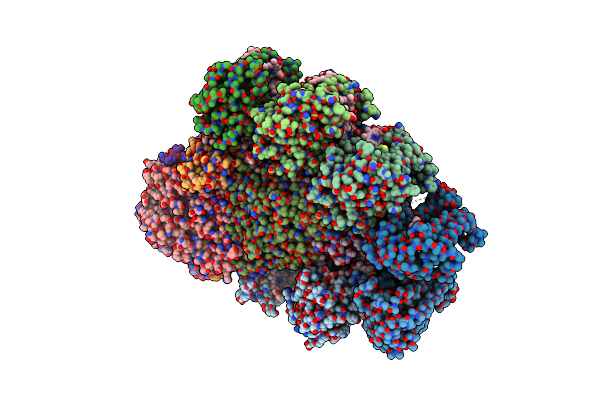 |
Organism: Nostoc sp.
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, LMT, LMG |
 |
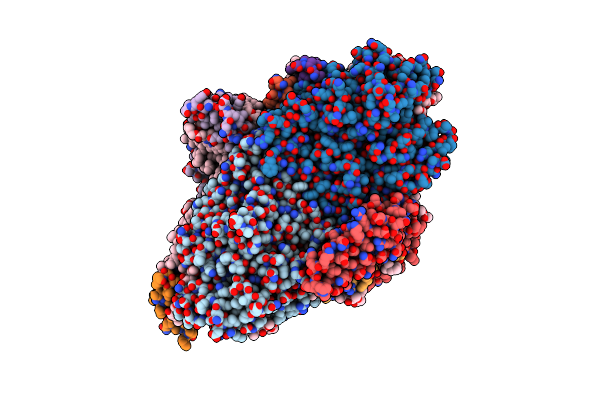
Organism: Gloeobacter violaceus (strain atcc 29082 / pcc 7421)
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, 1L3, SF4, BCR, LHG, LMG |
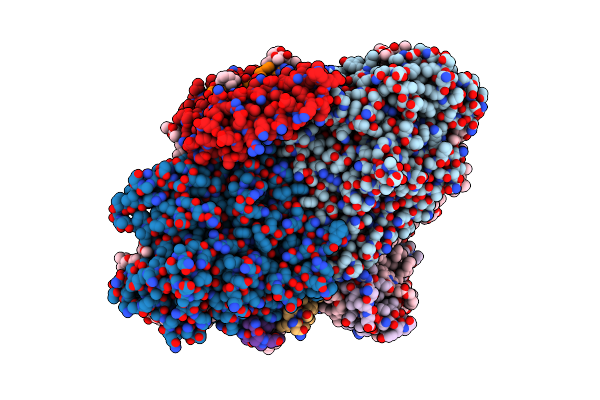
 |
Organism: Cyanophora paradoxa
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, LMG |
 |
Organism: Cyanophora paradoxa
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, LMG |
 |
Organism: Cyanophora paradoxa
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, BCR, LHG, SF4, LMG |
 |
Organism: Thermosynechococcus vulcanus
Method: ELECTRON MICROSCOPY Release Date: 2020-05-20 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, LMG, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-11-20 Classification: TRANSFERASE Ligands: C6F, SO4, GOL, CXS |
 |
Organism: Nostoc sp. (strain pcc 7120 / sag 25.82 / utex 2576)
Method: ELECTRON MICROSCOPY Release Date: 2019-11-13 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, LMG |
 |
Crystal Structure Analysis Of Cyanidioschyzon Melorae Ferredoxin D58N Mutant
Organism: Cyanidioschyzon merolae
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2013-08-07 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
Crystal Structure Of D(Cgcggatf5Ucgcg): 5-Formyluridine/Guanosine Base-Pair In B-Dna
|
 |
Crystal Structure Of D(Cgcggatf5Ucgcg): 5-Formyluridine:Guanosine Base-Pair In B-Dna With Hoechst33258
Method: X-RAY DIFFRACTION
Resolution:1.95 Å Release Date: 2011-04-27 Classification: DNA Ligands: HT, MG |
 |
Crystal Structure Of D(Cgcggatf5Ucgcg): 5-Formyluridine:Guanosine Base-Pair In B-Dna With Dapi
Method: X-RAY DIFFRACTION
Resolution:2.70 Å Release Date: 2011-04-27 Classification: DNA Ligands: DAP, MG |
 |
Crystal Structure Of An Alka Host/Guest Complex N7Methylguanine:Cytosine Base Pair
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-09-09 Classification: HYDROLASE/DNA |
 |
Crystal Structure Of D(Cgcgaatxcgcg) Where X Is 5-(N-Aminohexyl)Carbamoyl-2'-Deoxyuridine
Method: X-RAY DIFFRACTION
Resolution:1.55 Å Release Date: 2007-04-17 Classification: DNA Ligands: MG, CMY, K |
 |
Crystal Structure Of D(Cgcgaatxcgcg) Where X Is 5-(N-Aminohexyl)Carbamoyl-2'-Deoxyuridine
Method: X-RAY DIFFRACTION
Resolution:1.50 Å Release Date: 2007-04-17 Classification: DNA Ligands: CMY, K |
 |
Crystal Structure Of D(Cgcgaatxcgcg) Where X Is 5-(N-Aminohexyl)Carbamoyl-2'-O-Methyluridine
Method: X-RAY DIFFRACTION
Resolution:1.55 Å Release Date: 2007-04-17 Classification: DNA Ligands: CO, MG, CMY |
 |
Crystal Structure Of D(Cxctxcttc):R(Gaagaagag) Where X Is 5-(N-Aminohexyl)Carbamoyl-2'-O-Methyluridine
|
 |
Structural Analyses Of Dna:Dna And Rna:Dna Duplexes Containing 5-(N-Aminohexyl)Carbamoyl Modified Uridines
Method: X-RAY DIFFRACTION
Resolution:2.10 Å Release Date: 2007-04-17 Classification: DNA-RNA HYBRID Ligands: CMY |
 |
Crystal Structure Of D(Cxctxcttc):R(Gaagaagag) Where X Is 5-(N-Aminohexyl)Carbamoyl-2'-O-Methyluridine
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-04-17 Classification: DNA-RNA HYBRID Ligands: CMY |

