Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
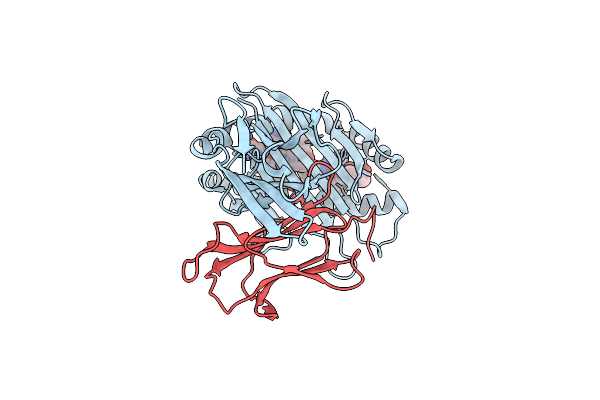 |
Complex Structure Of Hla-A*2402 With The Peptide From Hcov(Cov-2) Spike Protein
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-01-26 Classification: IMMUNE SYSTEM |
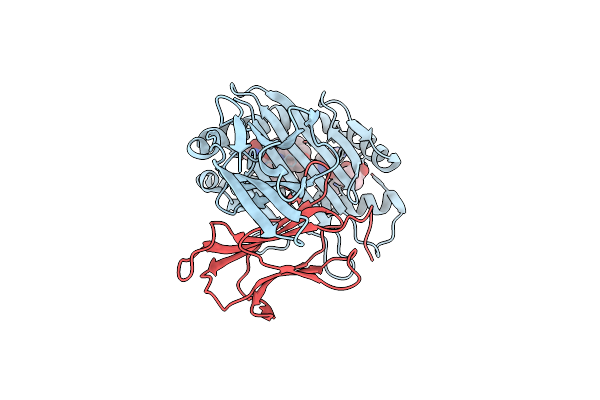 |
Complex Structure Of Hla-A*2402 With The Peptide From Hcov(Cov-229E) Spike Protein
Organism: Homo sapiens, Human coronavirus 229e
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-01-26 Classification: IMMUNE SYSTEM |
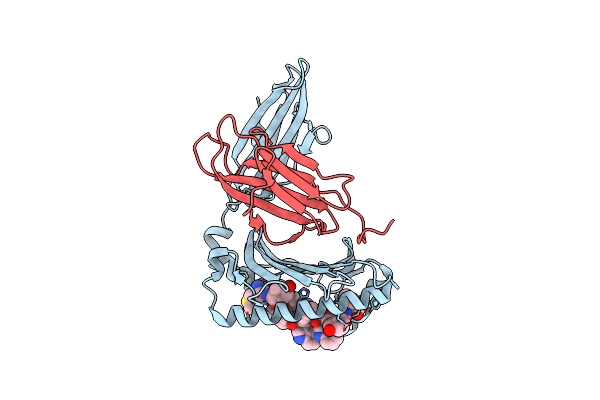 |
Complex Structure Of Hla-A*2402 With The Peptide From Hcov(Cov-Hku1) Spike Protein
Organism: Homo sapiens, Human coronavirus hku1
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-01-26 Classification: IMMUNE SYSTEM |
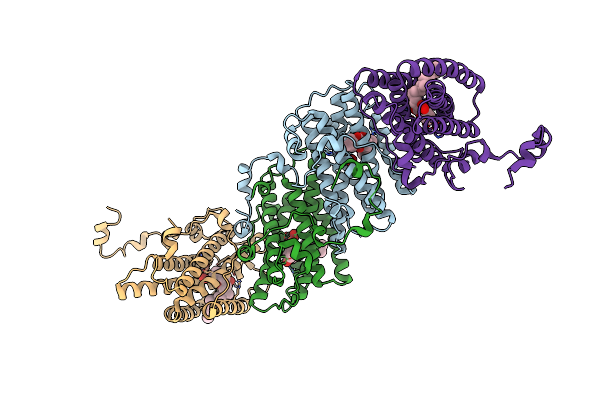 |
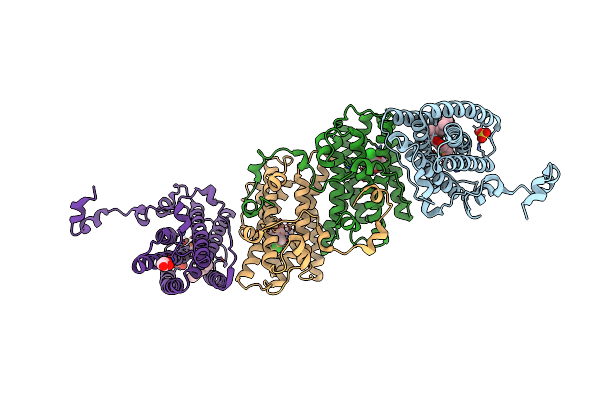
Crystal Structure Of Cyanide-Insensitive Alternative Oxidase From Trypanosoma Brucei With Ferulenol
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2019-02-27 Classification: MEMBRANE PROTEIN Ligands: FE, OH, OXY, 9AU |
 |
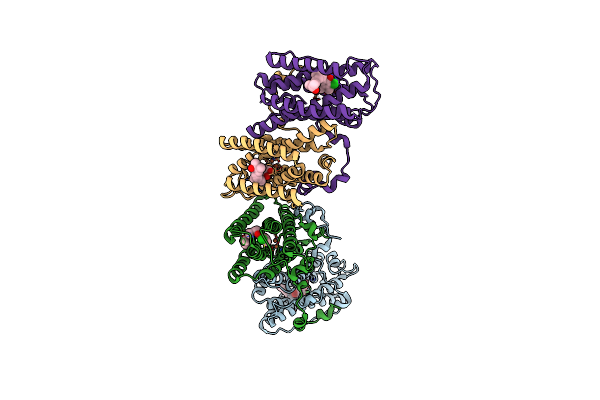
Crystal Structure Of Cyanide-Insensitive Alternative Oxidase From Trypanosoma Brucei With Colletochlorin B
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-02-27 Classification: MEMBRANE PROTEIN Ligands: FE, OH, RNB, SO4, GOL |
 |
Crystal Structure Of Cyanide-Insensitive Alternative Oxidase From Trypanosoma Brucei With Ascofuranone Derivative
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2019-02-27 Classification: MEMBRANE PROTEIN Ligands: FE, OH, CHW |
 |
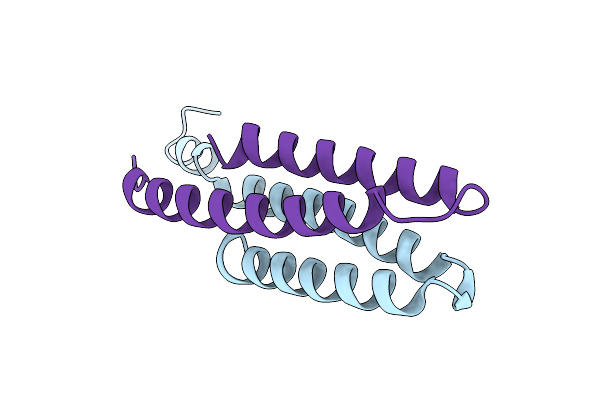
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2016-12-14 Classification: SIGNALING PROTEIN |
 |
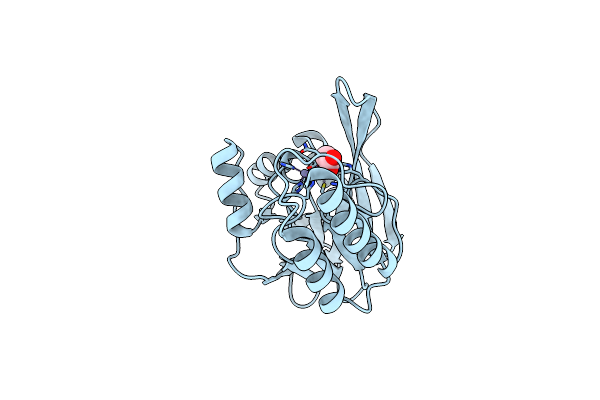
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-12-14 Classification: SIGNALING PROTEIN |
 |
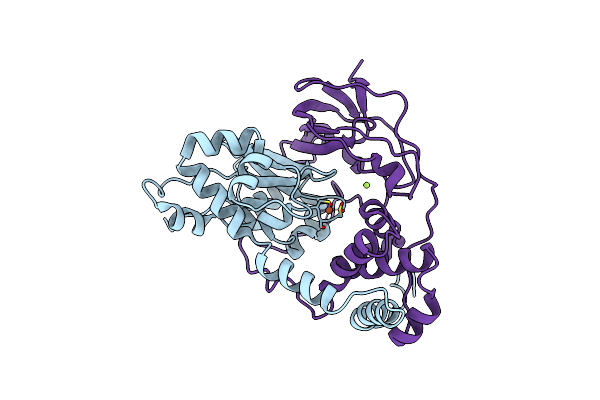
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-24 Classification: HYDROLASE Ligands: ZN, ACT |
 |
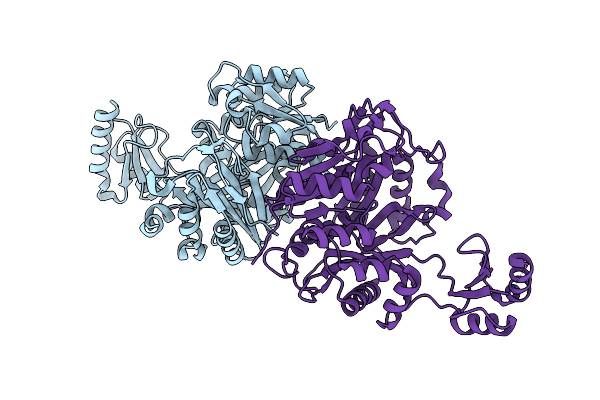
Mutational Study On Alpha-Gln90 Of Fe-Type Nitrile Hydratase From Rhodococcus Sp. N771
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Release Date: 2007-11-20 Classification: LYASE Ligands: FE, MG |
 |
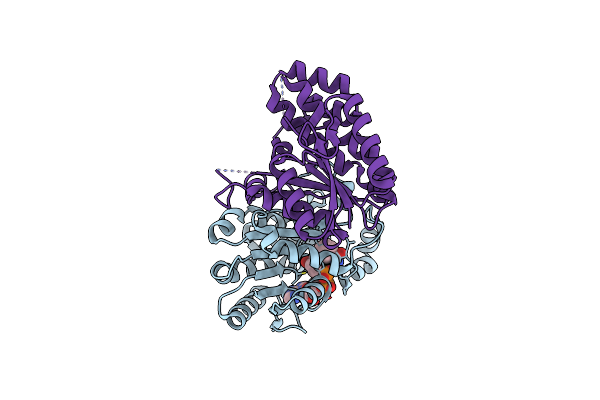
Organism: Geobacillus thermodenitrificans
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-09-25 Classification: LIGASE |
 |
Crystal Structure Of The 3-Alpha-Hydroxysteroid Dehydrogenase From Pseudomonas Sp. B-0831 Complexed With Nadh
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-08-15 Classification: OXIDOREDUCTASE Ligands: NAI |
 |
Crystal Structure Of The Carboxyl Transferase Subunit Of Putative Pcc Of Sulfolobus Tokodaii
Organism: Sulfolobus tokodaii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-07-04 Classification: LYASE |
 |
Structure-Based Design Of The Irreversible Inhibitors To Metallo--Lactamase (Imp-1)
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2005-06-21 Classification: HYDROLASE Ligands: ZN, OPS, ACY |
 |
Crystal Structure Of The Biotin Carboxylase Subunit Of Pyruvate Carboxylase
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-03-09 Classification: LIGASE |