Search Count: 273
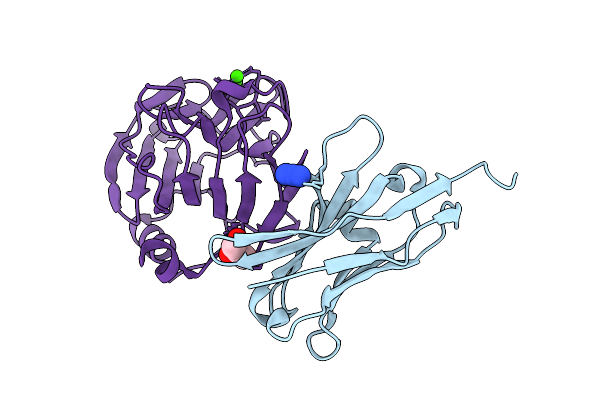 |
Crystal Structure Of The Complex Between Neuronal Pentraxin 2 (Np2 Ptx) And Antibody Fragment Vhh N1
Organism: Vicugna pacos, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PROTEIN BINDING Ligands: AZI, CA, CL, GOL |
 |
Organism: Homo sapiens
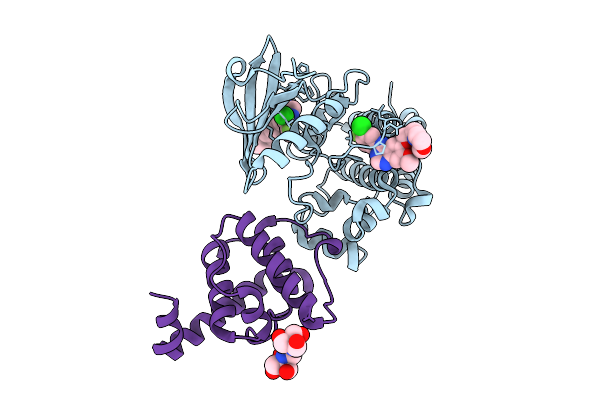
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE/INHIBITOR Ligands: IRE, B3P |
 |
Organism: Homo sapiens
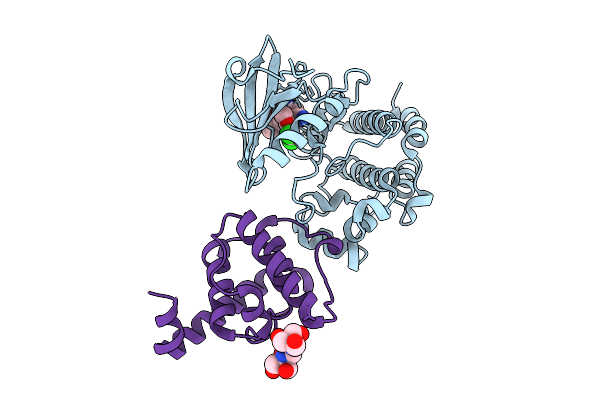
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE/INHIBITOR Ligands: DB8, B3P |
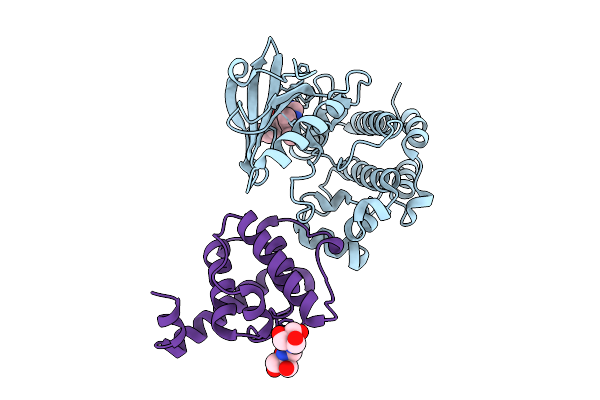 |
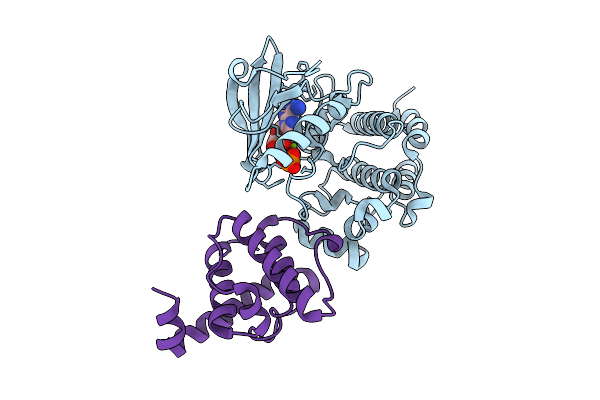
Crystal Structure Of The Ilk/Alpha-Parvin Core Complex Bound To 4-Methyl Erlotinib
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE/INHIBITOR Ligands: A1A2V, B3P |
 |
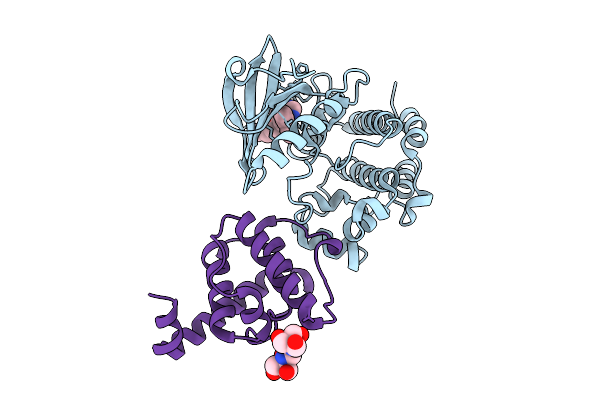
Crystal Structure Of The Ilk Mutant (R225A)/Alpha-Parvin Core Complex Bound To Mgatp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE/INHIBITOR Ligands: MG, ATP |
 |
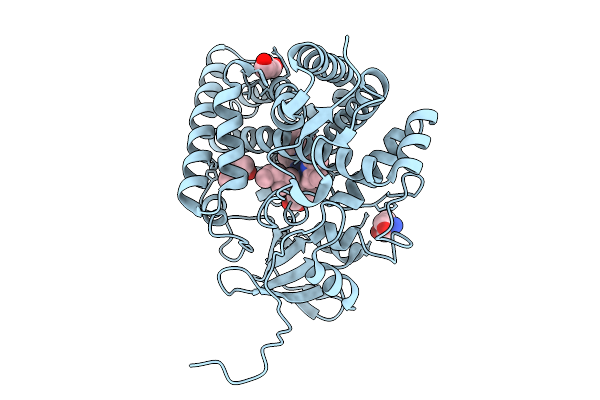
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE/INHIBITOR Ligands: AQ4, B3P |
 |
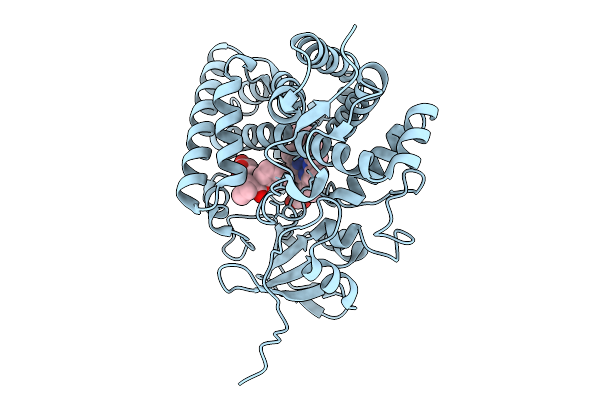
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM, MPD, TRS, PEG |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: HEM, 114 |
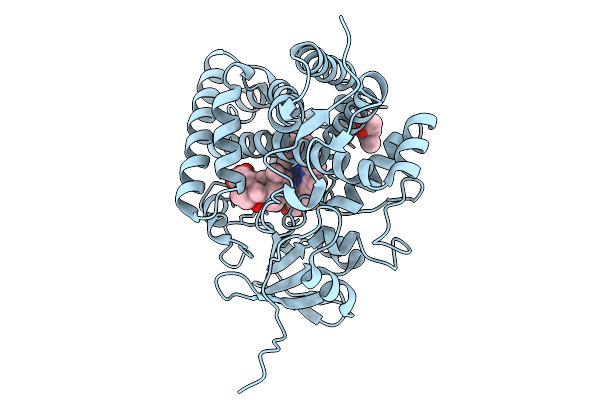 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: HEM, SIM, MPD |
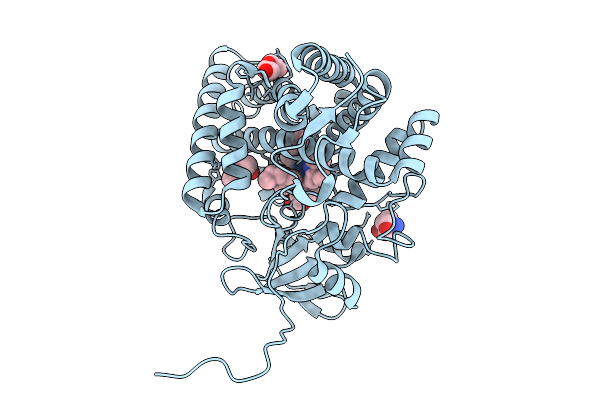 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: HEM, MPD, TRS, PEG |
 |
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN Ligands: ADP, MG |
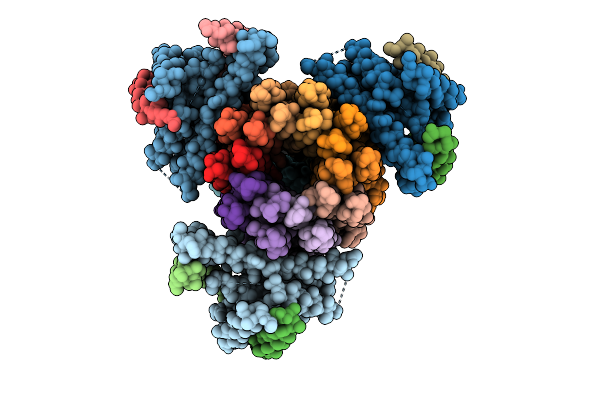 |
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN |
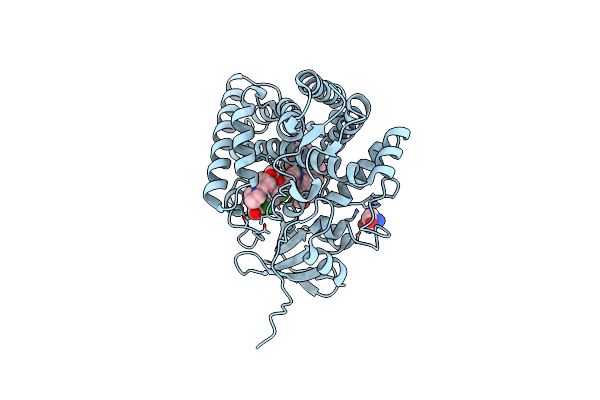 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: HEM, KTN, TRS |
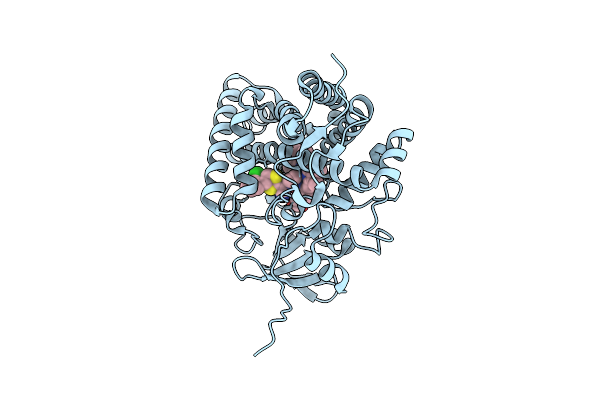 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: HEM, A1L6Q |
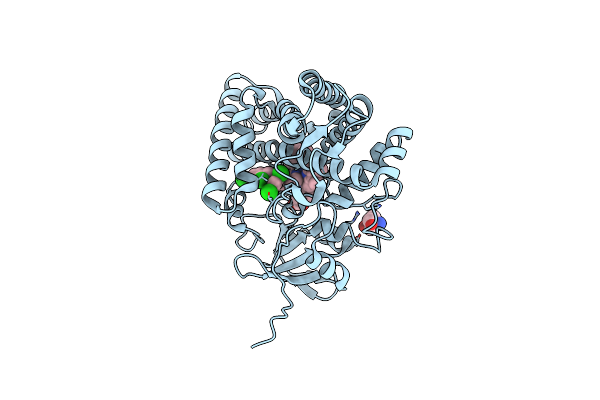 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: HEM, A1L6X, TRS |
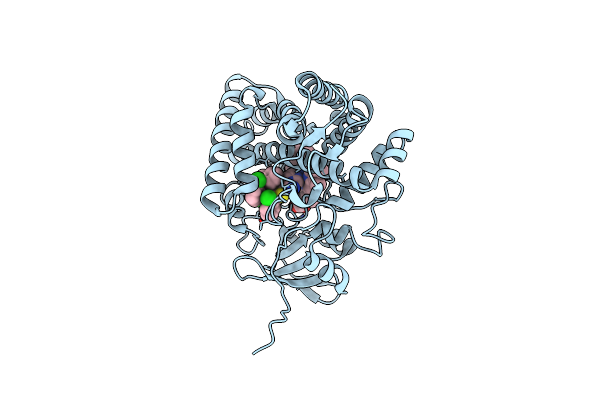 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: HEM, DIF, A1L6Q |
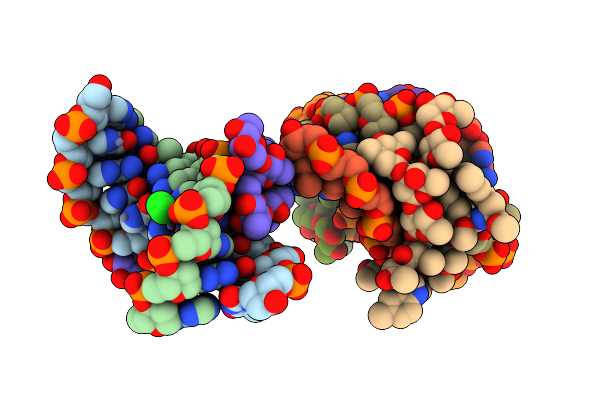 |
Crystal Structure Of Mithramycin Analogue Mtm Sa-5-Methyl-Trp In Complex With Dna Agaggcctct
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: DNA Ligands: ZN, A1BNI, CL, NA |
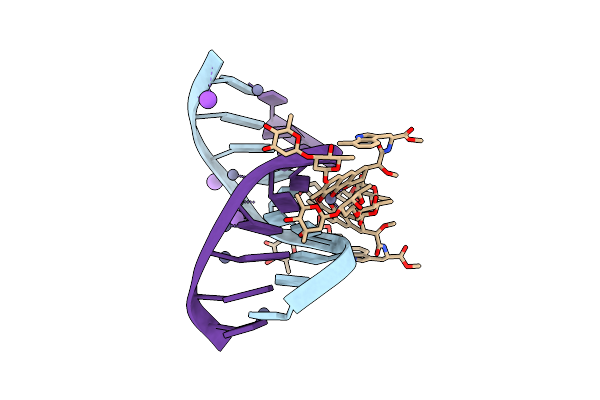 |
Crystal Structure Of Mithramycin Analogue Mtm Sa-7-Methyl-Trp In Complex With Double-Stranded Dna Agaggcctct
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: DNA Ligands: A1BNK, ZN, NA |
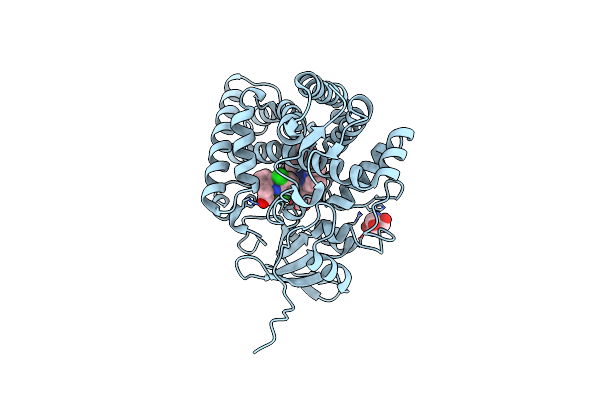 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-04-23 Classification: OXIDOREDUCTASE Ligands: HEM, DIF, TRS |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-04-23 Classification: OXIDOREDUCTASE Ligands: HEM, TRS, FLF, EDO |

