Search Count: 101
 |
Crystal Structure Of Cyp46A1 With [(1R,5S)-3-Oxa-8-Azabicyclo[3.2.1]Octan-8-Yl][(4R,8M)-8-(1,3-Oxazol-5-Yl)-6-(Trifluoromethyl)Imidazo[1,2-A]Pyridin-3-Yl]Methanone (Compound 3K)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE/INHIBITOR Ligands: HEM, A1BZE |
 |
Crystal Structure Of Cyp46A1 With (Morpholin-4-Yl)[(4R,8M)-8-(1,3-Oxazol-5-Yl)-6-(Trifluoromethyl)Imidazo[1,2-A]Pyridin-3-Yl]Methanone (Compound 2H)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE/INHIBITOR Ligands: HEM, A1BZD |
 |
Crystal Structure Of Cyp46A1 With Cyclopropyl[(4M)-4-(1,3-Oxazol-5-Yl)-6-(Trifluoromethyl)-1H-Indol-1-Yl]Methanone (Compound 2B)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE/INHIBITOR Ligands: HEM, A1BZC, EDO |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-10-09 Classification: TRANSFERASE Ligands: PLS, A1L3N |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2024-10-09 Classification: TRANSFERASE Ligands: PGE |
 |
Crystal Structure Of Human Methylmalonyl-Coa Mutase (Mmut) In Complex With Methylmalonic Acidemia Type A Protein (Mmaa), Coenzyme A, And Gdp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2023-08-09 Classification: Isomerase/Hydrolase Ligands: GDP, MG, COA |
 |
Crystal Structure Of Human Methylmalonyl-Coa Mutase In Complex With Adp And Cob(Ii)Alamin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-07-12 Classification: ISOMERASE Ligands: B12, ADP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-07-12 Classification: ISOMERASE Ligands: B12, GOL |
 |
Complex Structure Of Serine Hydroxymethyltransferase From Enterococcus Faecium And Its Inhibitor
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: 5M5, PLP, CL |
 |
Crystal Structure Of E. Faecium Shmt In Complex With (+)-Shin-1 And Plp-Ser
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: 5M5, PLS |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: THH, PLG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-02-23 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: HEM, EDO, 04Y, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-02-23 Classification: HYDROLASE Ligands: HEM, 05D |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-11-24 Classification: TRANSFERASE Ligands: MG, K, ATP, GOL |
 |
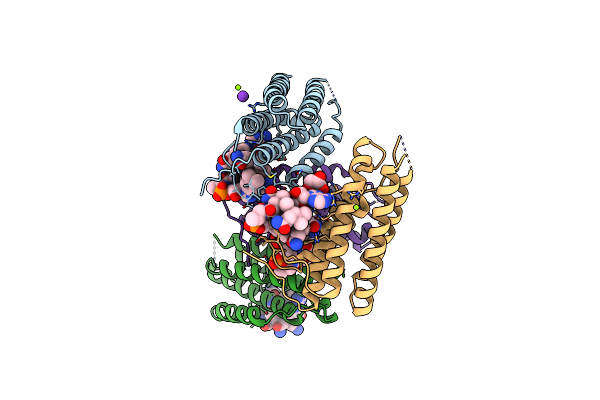
Structure Of Human Atp:Cobalamin Adenosyltransferase R190C Bound To Adenosylcobalamin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-11-24 Classification: TRANSFERASE Ligands: ACT, K, 5AD, B12 |
 |
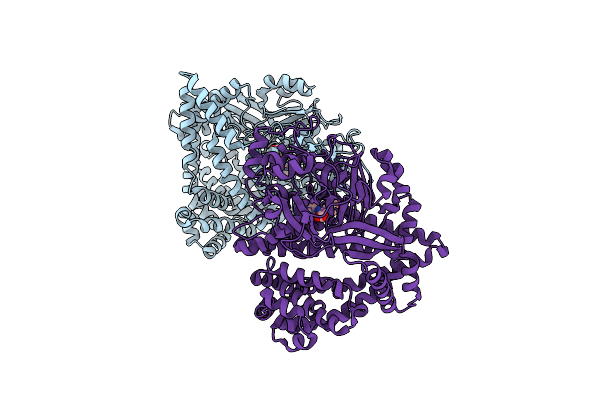
Structure Of Human Atp:Cobalamin Adenosyltransferase E193K Bound To Adenosylcobalamin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-11-24 Classification: TRANSFERASE Ligands: K, MG, B12, 5AD, GOL |
 |
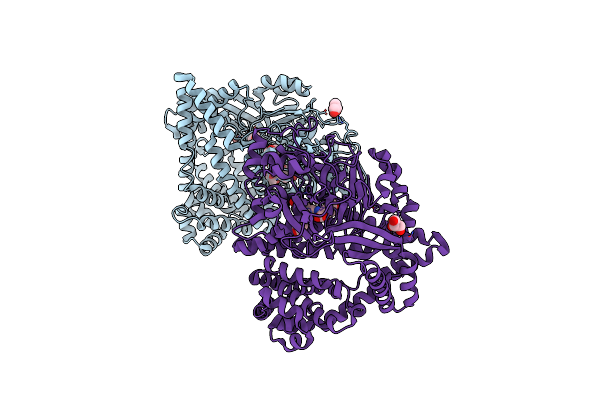
Organism: Stenotrophomonas maltophilia (strain r551-3)
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: VAL, TYR |
 |
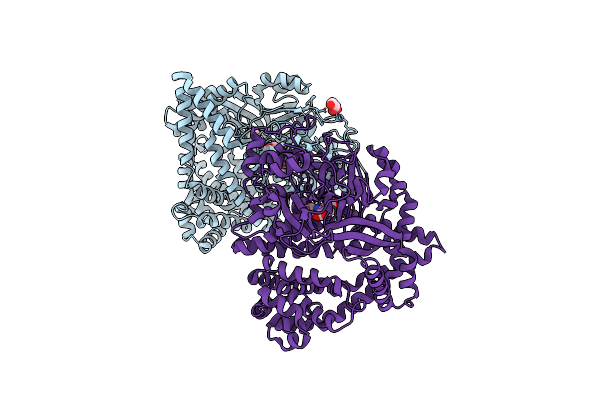
Organism: Stenotrophomonas maltophilia (strain r551-3)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: TYR, GOL |
 |
Organism: Stenotrophomonas maltophilia (strain r551-3)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: ASN, TYR, GOL |
 |
Organism: Stenotrophomonas maltophilia (strain r551-3)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: PHE, TYR, GOL |

