Search Count: 42
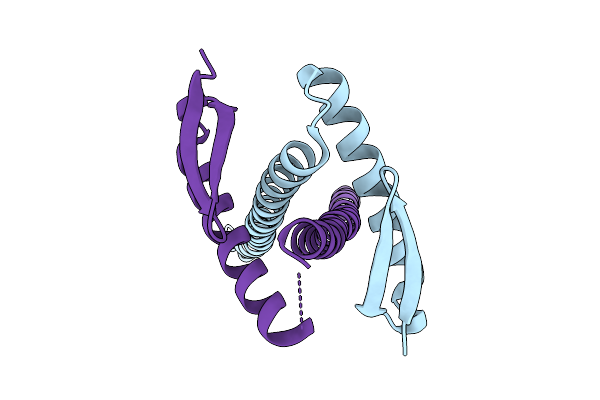 |
Organism: Klebsiella pneumoniae (strain 342)
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: CELL CYCLE Ligands: CL |
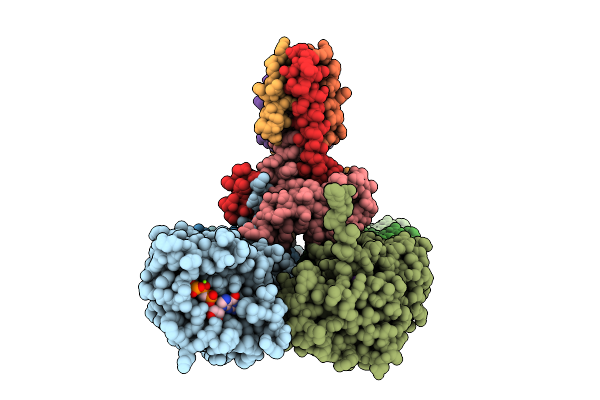 |
Organism: Klebsiella pneumoniae subsp. pneumoniae mgh 78578, Klebsiella pneumoniae 342
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: CELL CYCLE Ligands: G2P, MG, K |
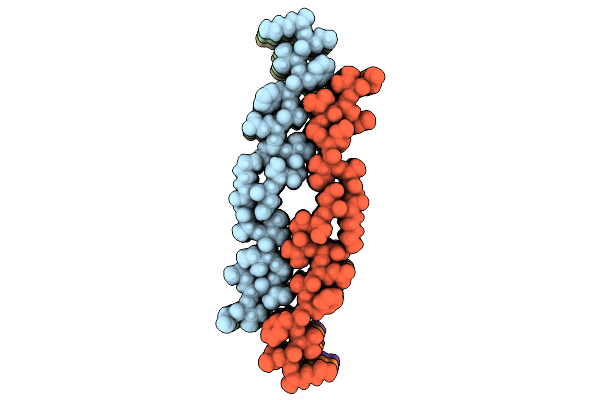 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: PROTEIN FIBRIL |
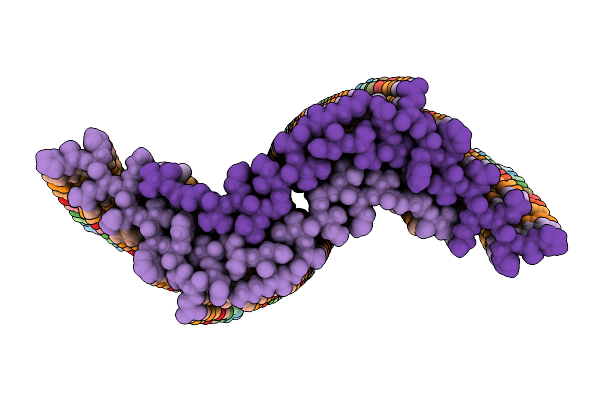 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: PROTEIN FIBRIL |
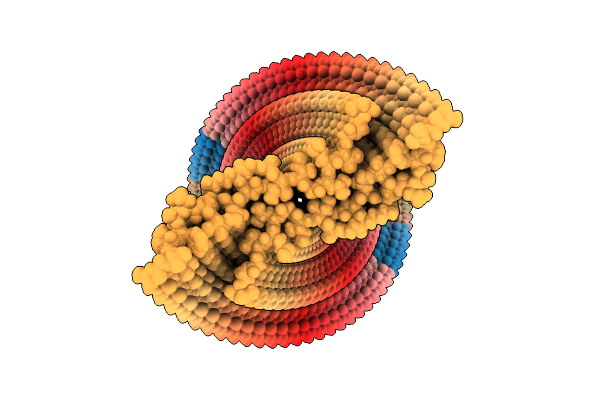 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: PROTEIN FIBRIL |
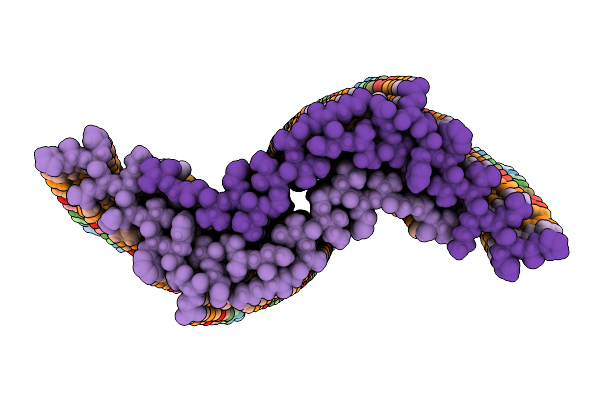 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: PROTEIN FIBRIL |
 |
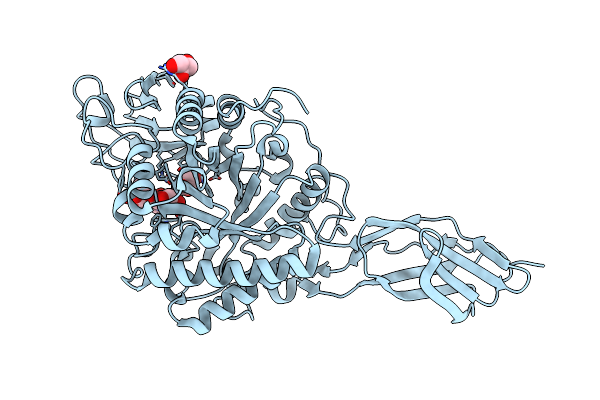
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-05-28 Classification: ELECTRON TRANSPORT Ligands: HEC, SO4, ACY, GOL, PEG |
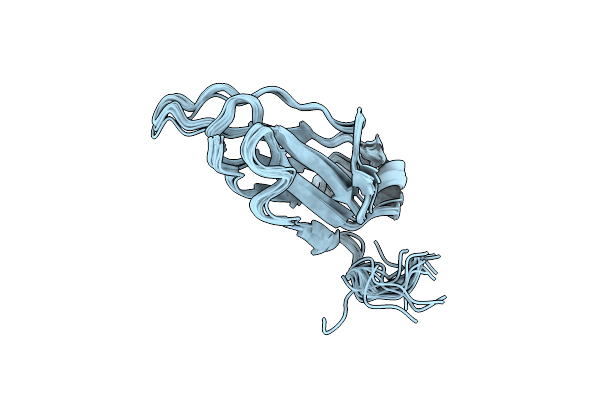 |
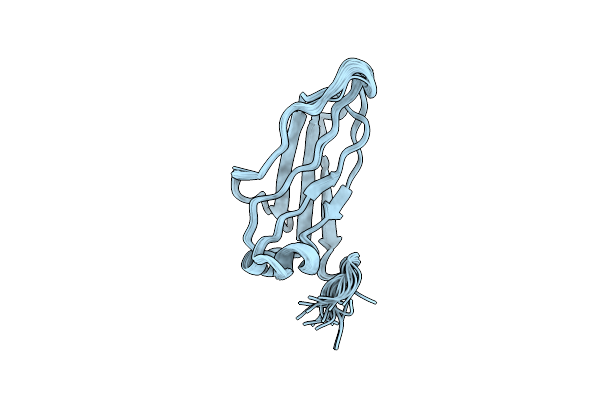 |
 |
 |
 |
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Guillardia theta ccmp2712
Method: ELECTRON MICROSCOPY Resolution:2.86 Å Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Organism: Guillardia theta ccmp2712
Method: ELECTRON MICROSCOPY Resolution:2.73 Å Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: RET, PC1 |
 |
Organism: Guillardia theta
Method: ELECTRON MICROSCOPY Resolution:2.71 Å Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: RET, PC1 |
 |
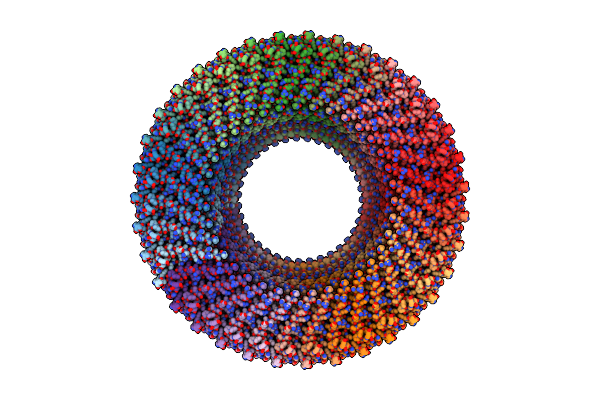
Structure Of The S-Ring Region Of The Vibrio Flagellar Ms-Ring Protein Flif With 34-Fold Symmetry Applied
Organism: Vibrio alginolyticus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: MOTOR PROTEIN |
 |
Structure Of The S-Ring Region Of The Vibrio Flagellar Ms-Ring Protein Flif With 35-Fold Symmetry Applied
Organism: Vibrio alginolyticus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: MOTOR PROTEIN |
 |
Organism: Mesocricetus auratus
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: GENE REGULATION Ligands: ADP, ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: GENE REGULATION Ligands: ADP, ATP |

