Search Count: 90
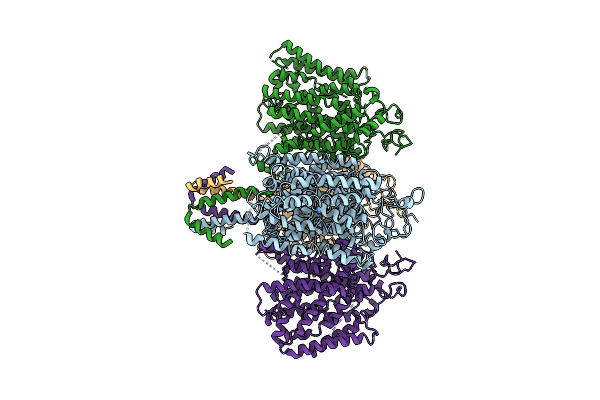 |
Organism: Zea mays
Method: ELECTRON MICROSCOPY Resolution:3.41 Å Release Date: 2025-11-05 Classification: CHOLINE-BINDING PROTEIN Ligands: URE |
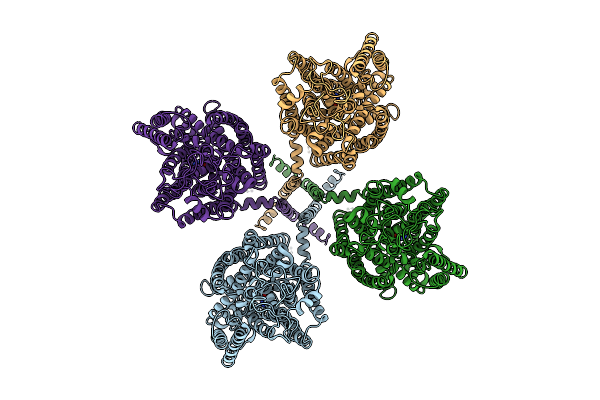 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.19 Å Release Date: 2025-11-05 Classification: PLANT PROTEIN Ligands: URE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: URE, PLM |
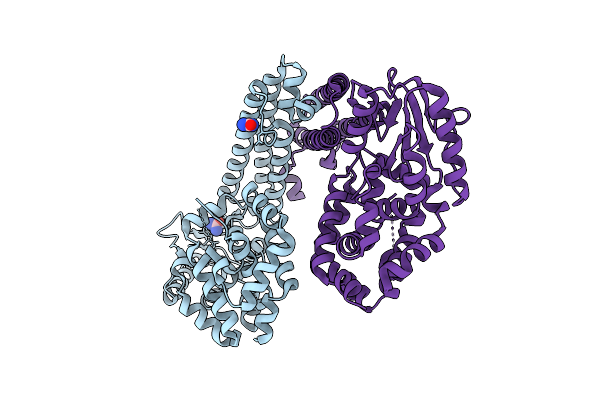 |
Organism: Gammaproteobacteria
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN Ligands: URE |
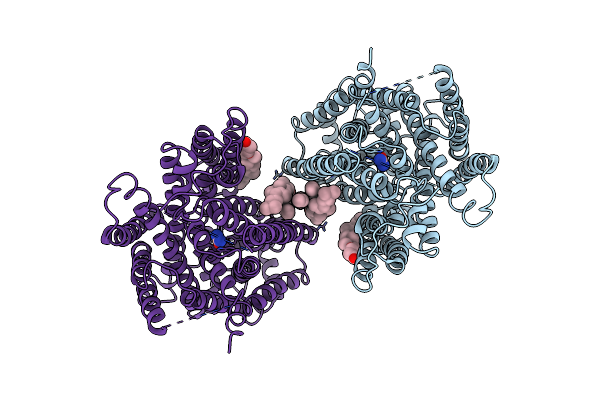 |
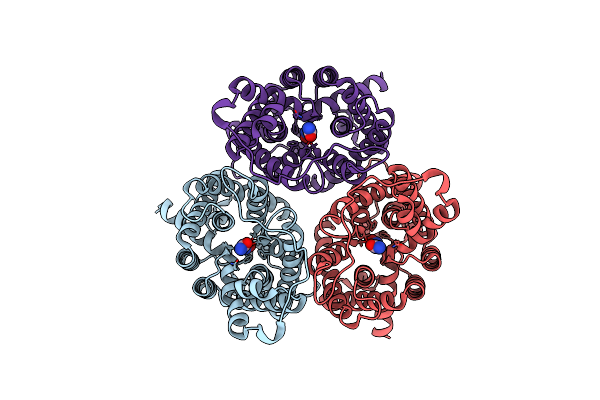
Arabidopsis High-Affinity Urea Transport Dur3 In The Urea-Bound Occluded Conformation, Dimeric State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: MEMBRANE PROTEIN Ligands: Y01, URE, R16, C14 |
 |
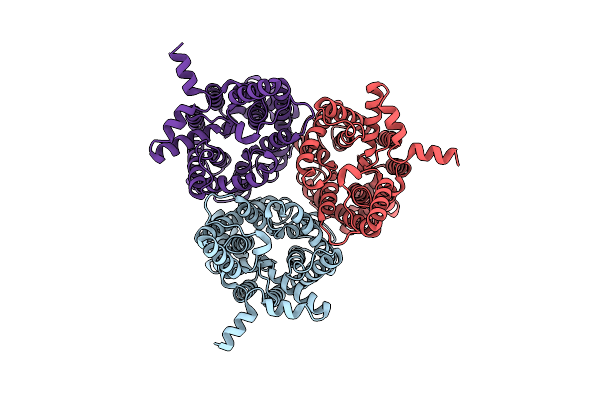
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: MEMBRANE PROTEIN Ligands: URE |
 |
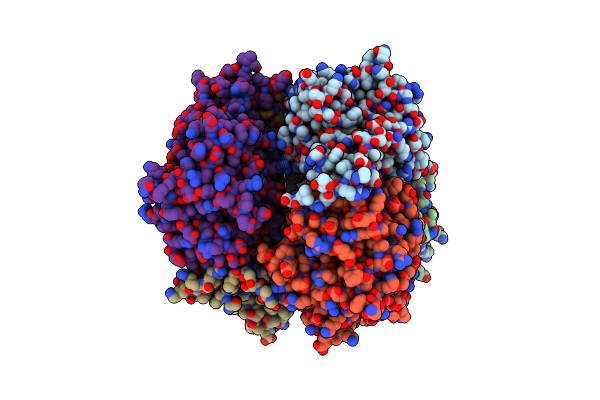
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: MEMBRANE PROTEIN Ligands: URE |
 |
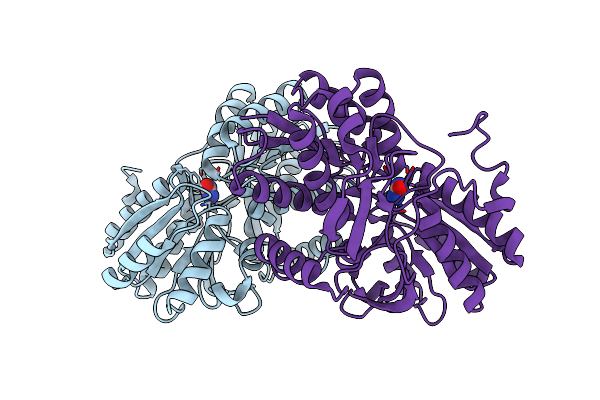
Metformin Hydrolase From Aminobacter Niigataensis Md1 With Urea In The Active Site
Organism: Aminobacter niigataensis
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2024-08-28 Classification: METAL BINDING PROTEIN Ligands: C5J, URE, NI, CA |
 |
Organism: Caldicellulosiruptor saccharolyticus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-06-19 Classification: TRANSPORT PROTEIN Ligands: URE, BR |
 |
The Structure Of Membrane-Active Antibiotic Cyclodecapeptide Gramicidin S In Complex With Urea
Organism: Brevibacillus brevis
Method: X-RAY DIFFRACTION Resolution:0.98 Å Release Date: 2024-03-06 Classification: ANTIBIOTIC Ligands: URE |
 |
Crystal Structure Of Urta From Prochlorococcus Marinus Str. Mit 9313 In Complex With Urea And Calcium
Organism: Prochlorococcus marinus str. mit 9313
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN Ligands: URE, CA |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: URE, CU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2022-11-16 Classification: ONCOPROTEIN Ligands: URE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2022-11-16 Classification: ONCOPROTEIN Ligands: URE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2022-11-16 Classification: ONCOPROTEIN Ligands: URE |
 |
Lysozyme Crystallized In The Presence Of The Hydrated Deep Eutectic Solvent Choline Chloride-Urea 1:2
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-10-20 Classification: HYDROLASE Ligands: ACT, CHT, CL, NA, URE |
 |
Crystal Structure Of Urta1 From Synechococcus Wh8102 In Complex With Urea And Calcium
Organism: Synechococcus sp. (strain wh8102)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-10-20 Classification: TRANSPORT PROTEIN Ligands: URE, EDO, CA |
 |
Crystal Structure Of Urta From Synechococcus Cc9311 In Complex With Urea And Calcium
Organism: Synechococcus sp. (strain cc9311)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-10-13 Classification: TRANSPORT PROTEIN Ligands: URE, EDO, CA, CL |
 |
Crystal Structure Of The C-Src Sh3 Domain Mutant V111L-N113S-T114S In 7 M Urea
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-08-25 Classification: PROTEIN BINDING Ligands: URE, NA |
 |
Crystal Structure Of The C-Src Sh3 Domain Mutant V111L-N113S-T114S In 3 M Urea
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2021-08-25 Classification: PROTEIN BINDING Ligands: URE |

