Search Count: 73
 |
Ligand Binding Domain Of Roseburia Intestinalis L1-82 Uracil Chemoreceptor (Dcache) In Complex With Uracil And Acetate
Organism: Roseburia intestinalis l1-82
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2025-03-12 Classification: SIGNALING PROTEIN Ligands: URA, ACT, EDO |
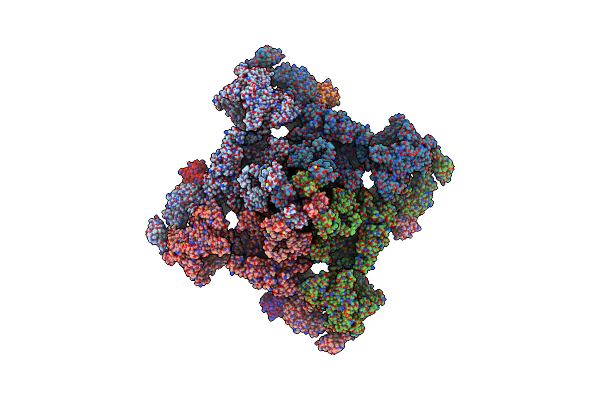 |
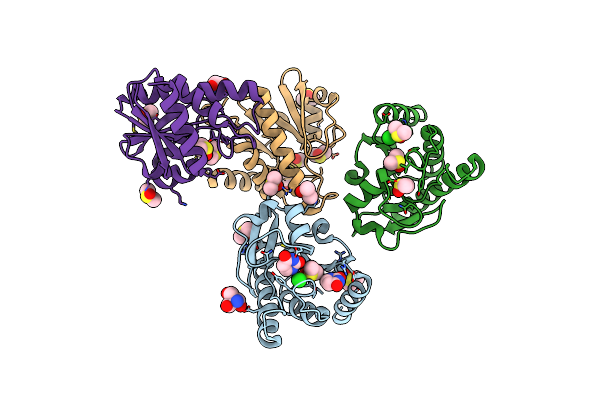
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ATP, CA, ZN, URA |
 |
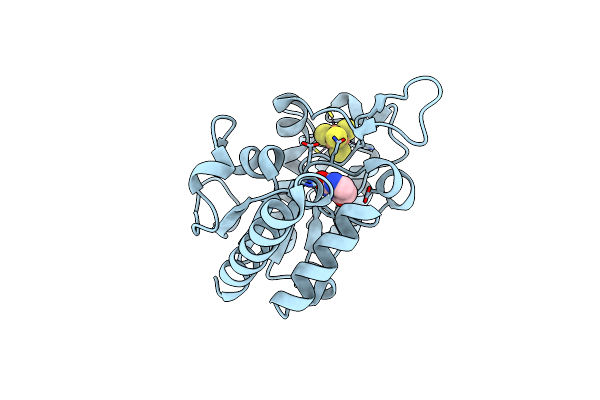
Group Deposition For Crystallographic Fragment Screening Of Chikungunya Virus Nsp3 Macrodomain -- Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain In Complex With Z56889474 (Chikv_Macb-X1365)
Organism: Chikungunya virus, Chikungunya virus
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2024-05-29 Classification: VIRAL PROTEIN Ligands: DMS, TRS, CL, URA |
 |
Wide Inward-Open Liganded Uraa In Complex With A Conformation-Selective Synthetic Nanobody
Organism: Escherichia coli o157:h7, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2024-04-10 Classification: MEMBRANE PROTEIN Ligands: 1PE, BNG, URA, DMU |
 |
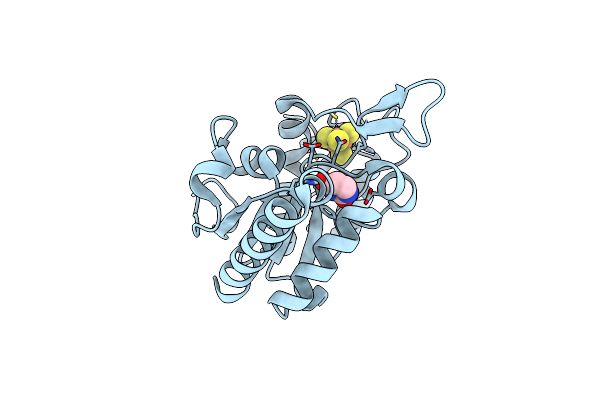
Complex Form Of Msmudgx And Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Msmudgx
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2023-06-21 Classification: DNA BINDING PROTEIN Ligands: SF4, URA |
 |
Complex Form Of Msmudgx H109A Mutant And Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-06-21 Classification: DNA BINDING PROTEIN Ligands: SF4, URA |
 |
Complex Form Of Msmudgx H109C Mutant And Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Msmudgx H109C
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-06-21 Classification: DNA BINDING PROTEIN Ligands: SF4, URA, GOL |
 |
Complex Form Of Msmudgx H109G Mutant And Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Msmudgx H109G
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-06-21 Classification: DNA BINDING PROTEIN Ligands: SF4, URA |
 |
Complex Form Of Msmudgx H109K Mutant And Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Msmudgx H109K
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-06-21 Classification: DNA BINDING PROTEIN Ligands: SF4, URA |
 |
Complex Form Of Msmudgx H109Q Mutant And Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Msmudgx H109Q
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2023-06-21 Classification: DNA BINDING PROTEIN Ligands: SF4, URA, BME |
 |
Complex Form Of Msmudgx H109S/R184A Double Mutant And Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Msmudgx H109S/R184A
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-06-21 Classification: DNA BINDING PROTEIN Ligands: SF4, URA |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-06-01 Classification: TRANSCRIPTION Ligands: URA, EDO |
 |
Organism: Shewanella oneidensis mr-1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: SO4, GOL, URA |
 |
Dihydropyrimidine Dehydrogenase (Dpd) C671A Mutant Soaked With Uracil And Nadph Anaerobically
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-06-09 Classification: FLAVOPROTEIN, Oxidoreductase Ligands: SF4, FAD, URA, FNR, ALA, LEU, PRO |
 |
Crystal Structure Of Monooxygenase Ruta Complexed With Uracil Under Atmospheric Pressure.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-02-05 Classification: FLAVOPROTEIN Ligands: FMN, URA, SO4 |
 |
Crystal Structure Of Monooxygenase Ruta Complexed With Uracil And Dioxygen Under 1.5 Mpa / 15 Bars Of Oxygen Pressure.
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-05 Classification: FLAVOPROTEIN Ligands: FMN, URA, SO4, OXY |
 |
Crystal Structure Of A Fragment Of E. Coli Trna(Asp) Consisting Of Its Acceptor Stem/T Stem-Loop. Short Unit Cell.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-01-01 Classification: RNA Ligands: SO4, URA, DPO |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-07-24 Classification: HYDROLASE Ligands: SF4, URA, SO4 |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: URA, SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2019-07-24 Classification: HYDROLASE Ligands: SF4, URA |

