Search Count: 21
 |
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.76 Å Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: UGA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TRANSFERASE Ligands: MN, UGA |
 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: 3PE, NAG, UGA, Y01, MN |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2023-08-02 Classification: OXIDOREDUCTASE Ligands: UGA |
 |
Crystal Structure Of Full-Length Human Lysyl Hydroxylase Lh3 - Cocrystal With Fe2+, Mn2+, Udp-Glucuronic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2021-05-19 Classification: TRANSFERASE Ligands: AKG, FE2, MN, UGA, NAG, EDO |
 |
Crystal Structure Of Full-Length Human Lysyl Hydroxylase Lh3 - Val80Lys Mutant - Cocrystal With Fe2+, Mn2+, Udp-Glucuronic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-05-19 Classification: TRANSFERASE Ligands: UGA, GOL, AKG, FE2, MN, NAG |
 |
Crystal Structure Of Udp-Glucuronic Acid 4-Epimerase From Bacillus Cereus In Complex With Udp-Glucuronic Acid And Nad
Organism: Bacillus cereus hua2-4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-07-29 Classification: OXIDOREDUCTASE Ligands: NAD, UGA |
 |
Equilibrium Structure Of Udp-Glucuronic Acid 4-Epimerase From Bacillus Cereus In Complex With Udp-Glucuronic Acid/Udp-Galacturonic Acid And Nad
Organism: Bacillus cereus hua2-4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-07-29 Classification: OXIDOREDUCTASE Ligands: NAD, UGA, UGB |
 |
Organism: Streptomyces viridosporus atcc 14672
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2019-11-13 Classification: BIOSYNTHETIC PROTEIN Ligands: NAD, UGA |
 |
The Structure Of C100A Mutant Of Arabidopsis Thaliana Udp-Apiose/Udp-Xylose Synthase In Complex With Nadh And Udp-D-Glucuronic Acid
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.47 Å Release Date: 2019-10-23 Classification: OXIDOREDUCTASE Ligands: NAD, UGA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2013-08-07 Classification: LYASE Ligands: NAD, UDP, UGA, SO4, POP |
 |
Crystal Structure Of Udp-Glucose Dehydrogenase From Klebsiella Pneumoniae Complexed With Product Udp-Glucuronic Acid
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-09-28 Classification: OXIDOREDUCTASE Ligands: CXS, UGA |
 |
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-07-27 Classification: OXIDOREDUCTASE Ligands: UGA, SO4, ACT, GOL, TRS |
 |
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-07-27 Classification: OXIDOREDUCTASE Ligands: UGA, SO4 |
 |
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-07-27 Classification: OXIDOREDUCTASE Ligands: UGA, SO4, ACT, GOL |
 |
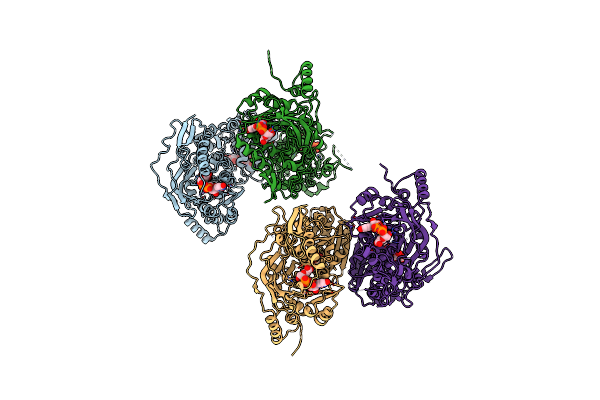
Crystal Structure Of Udp-Glucose 6-Dehydrogenase From Porphyromonas Gingivalis Bound To Product Udp-Glucuronate
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-03-24 Classification: OXIDOREDUCTASE Ligands: UGA |
 |
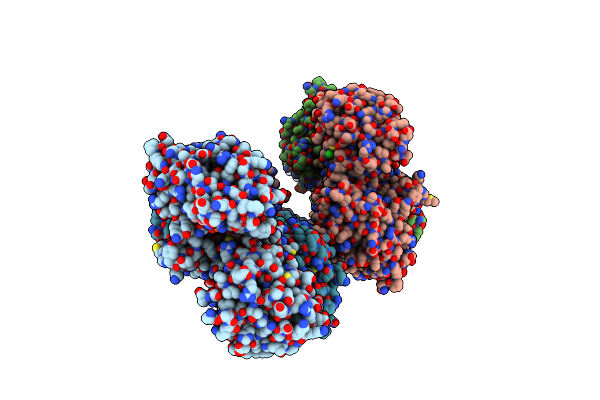
Crystal Structure Of Chondroitin Polymerase From Escherichia Coli Strain K4 (K4Cp) Complexed With Udp-Glcua And Udp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-09-16 Classification: TRANSFERASE Ligands: UGA, MN, UDP |
 |
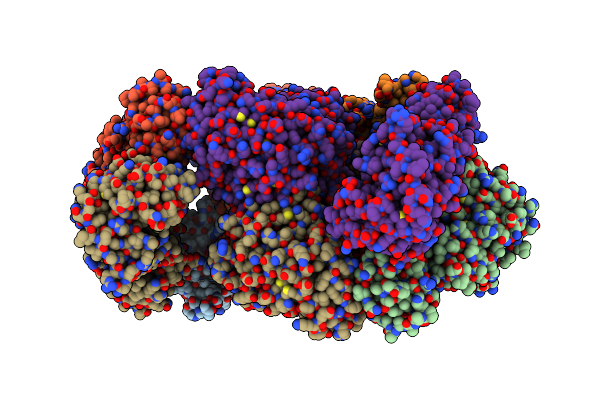
Crystal Structure Of Human Udp-Glucose Dehydrogenase Product Complex With Udp-Glucuronate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-07-10 Classification: OXIDOREDUCTASE Ligands: NAD, UGA, CL, EDO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2005-06-07 Classification: HYDROLASE Ligands: ATP, UGA |
 |
Crystal Structure Of Beta1,3-Glucuronyltransferase I In Complex With The Active Udp-Glcua Donor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2002-06-19 Classification: TRANSFERASE Ligands: MN, UGA |

