Search Count: 78
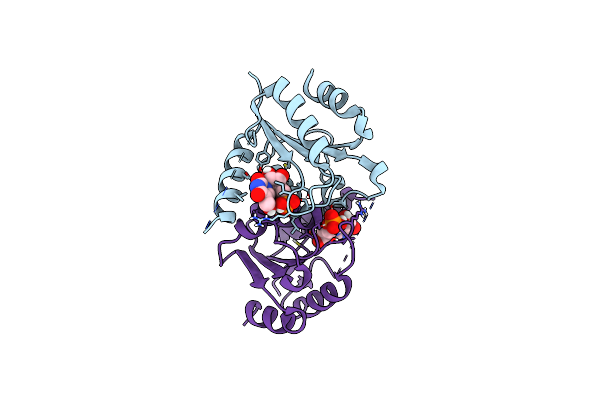 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBB |
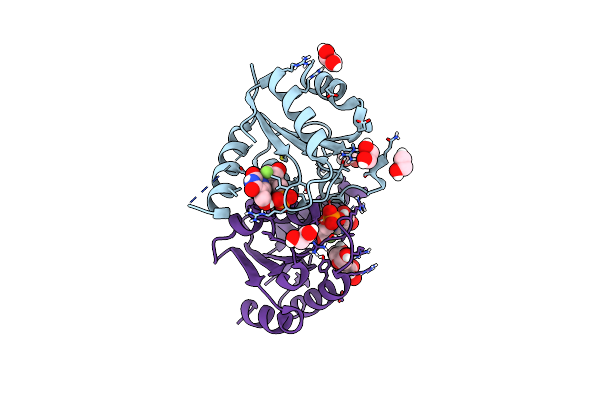 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBC, EDO, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBE |
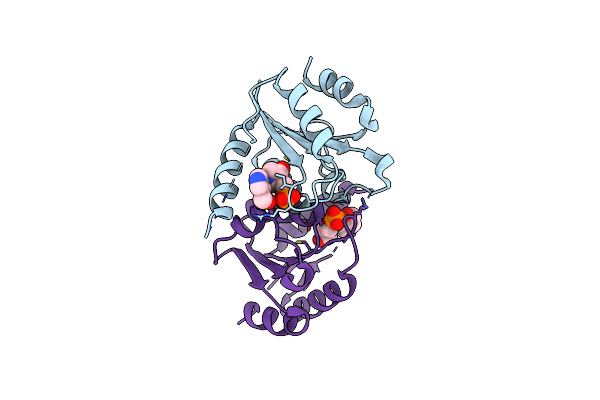 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: A1BBF |
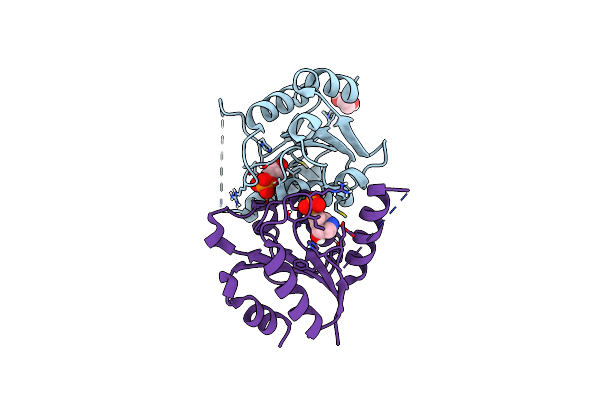 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-02-05 Classification: HYDROLASE/INHIBITOR Ligands: NR1, EDO |
 |
Crystal Structure Of The Trypanosoma Cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase (Hgxprt), Isoform D, Bound To The Dead-End Complex Xanthine And Pyrophosphate
Organism: Trypanosoma cruzi (strain cl brener)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2023-07-19 Classification: TRANSFERASE Ligands: XAN, PO4 |
 |
Crystal Structure Of The Trypanosoma Cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase (Hgxprt), Isoform D, Bound To Hydroxypropyl-Lin-Immh Phosphonate
Organism: Trypanosoma cruzi (strain cl brener)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-07-19 Classification: TRANSFERASE/INHIBITOR Ligands: YCE, PO4 |
 |
Crystal Structure Of The Trypanosoma Cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase (Hgxprt), Isoform D, Bound To (S)-Serme-Immh Phosphonate
Organism: Trypanosoma cruzi (strain cl brener)
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2023-07-19 Classification: TRANSFERASE/INHIBITOR Ligands: YC9 |
 |
Crystal Structure Of The Trypanosoma Cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase (Hgxprt), Isoform D, Bound To (R)-Serme-Immh Phosphonate
Organism: Trypanosoma cruzi (strain cl brener)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-07-19 Classification: TRANSFERASE/INHIBITOR Ligands: YC9 |
 |
Crystal Structure Of The Trypanosoma Cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase (Hgxprt), Isoform D, Bound To Immucillin-Hp
Organism: Trypanosoma cruzi (strain cl brener)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2023-07-19 Classification: TRANSFERASE Ligands: IRP |
 |
Crystal Structure Of The Trypanosoma Cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase (Hgxprt), Isoform D, Bound To Immucillin-Gp, Showing The Structure Of The Complete Active Site In Its Open Conformation
Organism: Trypanosoma cruzi (strain cl brener)
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2023-07-19 Classification: TRANSFERASE/INHIBITOR Ligands: IMU |
 |
Crystal Structure Of Plasmodium Falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase In Complex With [(3S)-4-Hydroxy-3-[({2-Amino-4-Hydroxy-5H-Pyrrolo[3,2-D]Pyrimidin-7-Yl}Methyl)Amino]Butyl]Phosphonic Acid
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-12-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: YBM, POP, MG, EDO, ACY |
 |
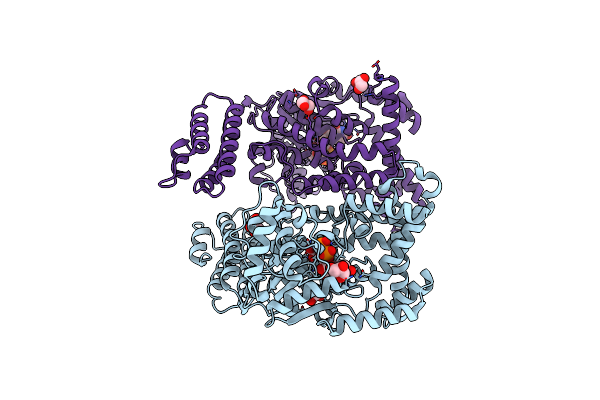
Crystal Structure Of Clostridium Difficile Toxin B (Tcdb) Glucosyltransferase In Complex With Udp And Isofagomine
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-11-17 Classification: TRANSFERASE/Inhibitor Ligands: EDO, UDP, IFM, MN |
 |
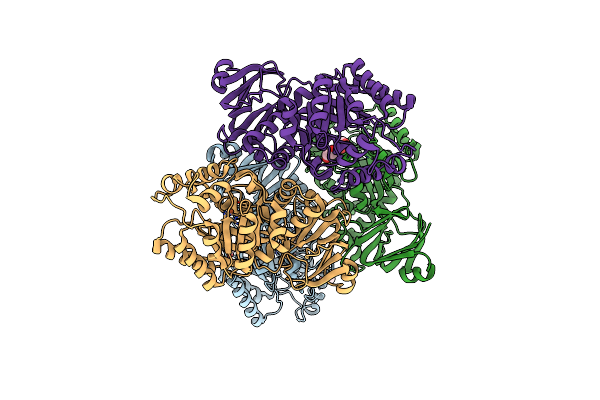
Crystal Structure Of Clostridium Difficile Toxin B (Tcdb) Glucosyltransferase In Complex With Udp And Noeuromycin
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-11-17 Classification: TRANSFERASE/Inhibitor Ligands: NOY, UDP, GOL, MN |
 |
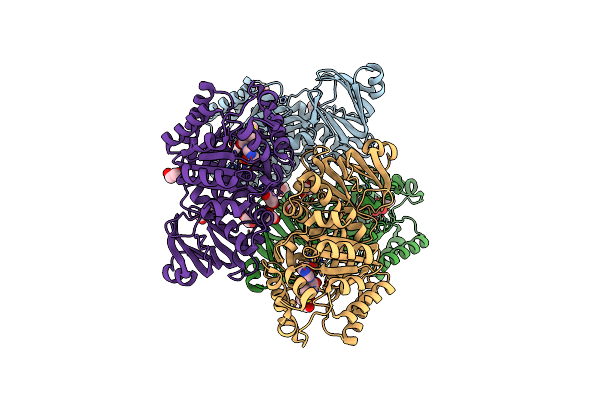
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2021-06-23 Classification: HYDROLASE Ligands: FE, EDO |
 |
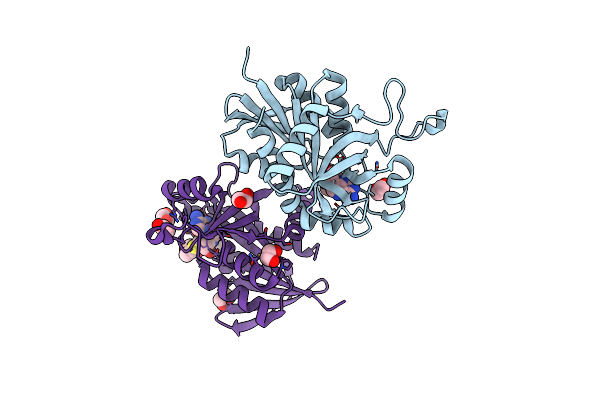
Crystal Structure Of Helicobacter Pylori Aminofutalosine Deaminase (Aflda) In Complex With Methylthio-Coformycin
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2021-06-23 Classification: HYDROLASE/INHIBITOR Ligands: FE, EDO, MCF |
 |
Crystal Structure Of Helicobacter Pylori 5'-Methylthioadenosine/S-Adenosyl Homocysteine Nucleosidase (Mtan) Complexed With (3R,4S)-1-((4-Amino-5H-Pyrrolo[3,2-D]Pyrimidin-7-Yl)Methyl)-4-((Prop-2-Yn-1-Ylthio)Methyl)Pyrrolidin-3-Ol
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-03-20 Classification: HYDROLASE Ligands: EDO, OS2, NH4 |
 |
Crystal Structure Of Helicobacter Pylori 5'-Methylthioadenosine/S-Adenosyl Homocysteine Nucleosidase (Mtan) Complexed With (3R,4S)-1-((4-Amino-5H-Pyrrolo[3,2-D]Pyrimidin-7-Yl)Methyl)-4-((Pent-4-Yn-1-Ylthio)Methyl)Pyrrolidin-3-Ol
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2019-03-20 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: EDO, OS3 |
 |
Crystal Structure Of Helicobacter Pylori 5'-Methylthioadenosine/S-Adenosyl Homocysteine Nucleosidase (Mtan) Complexed With (3R,4S)-1-((4-Amino-5H-Pyrrolo[3,2-D]Pyrimidin-7-Yl)Methyl)-4-(((3-(1-Benzyl-1H-1,2,3-Triazol-4-Yl)Propyl)Thio)Methyl)Pyrrolidin-3-Ol
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-03-20 Classification: HYDROLASE Ligands: EDO, OS5, NH4 |

