Search Count: 6
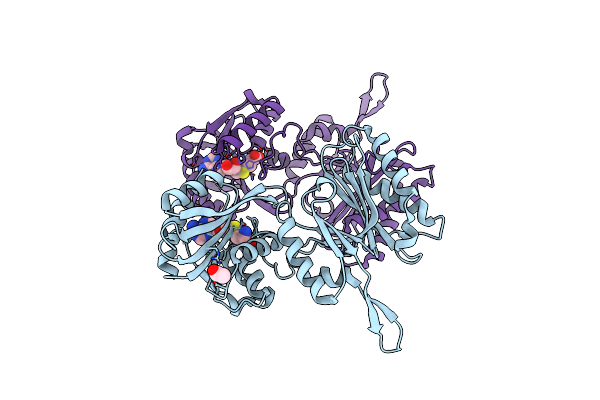 |
Crystal Structure Of Trm14 From Pyrococcus Furiosus In Complex With S-Adenosyl-L-Homocysteine
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-03-14 Classification: TRANSFERASE Ligands: SAH, ACT |
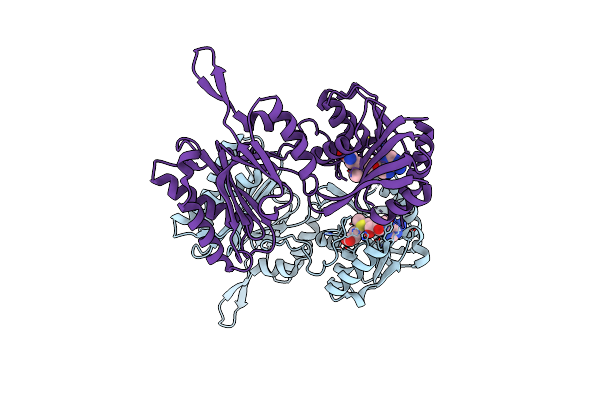 |
Crystal Structure Of Trm14 From Pyrococcus Furiosus In Complex With S-Adenosylmethionine
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-03-14 Classification: TRANSFERASE Ligands: SAM |
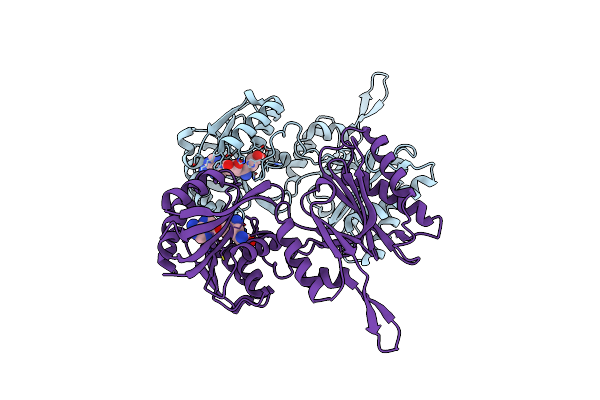 |
Crystal Structure Of Trm14 From Pyrococcus Furiosus In Complex With Sinefungin
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2012-03-14 Classification: TRANSFERASE Ligands: SFG |
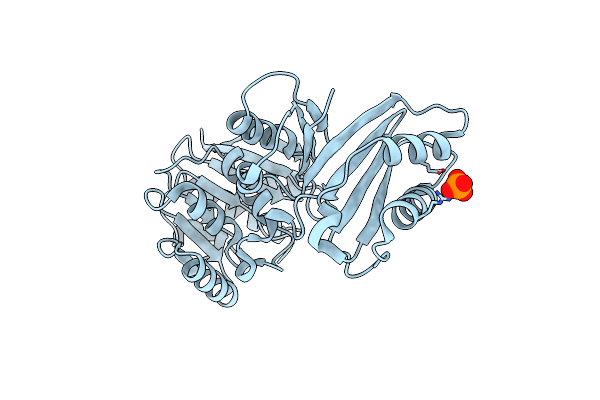 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2012-03-14 Classification: TRANSFERASE Ligands: PO4 |
 |
Atomic Model Of The Ocr-Bound Methylase Complex From The Type I Restriction-Modification Enzyme Ecoki (M2S1). Based On Fitting Into Em Map 1534.
Organism: Enterobacteria phage t7, Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:18.00 Å Release Date: 2011-02-09 Classification: TRANSFERASE |
 |
Atomic Model Of The Dna-Bound Methylase Complex From The Type I Restriction-Modification Enzyme Ecoki (M2S1). Based On Fitting Into Em Map 1534.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:18.00 Å Release Date: 2011-02-09 Classification: TRANSFERASE/DNA Ligands: SAM |

