Search Count: 86
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-10-01 Classification: IMMUNE SYSTEM |
 |
Organism: Escherichia coli uti89, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: CELL ADHESION |
 |
Organism: Escherichia coli uti89, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: CELL ADHESION |
 |
Organism: Escherichia coli uti89, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: CELL ADHESION |
 |
Organism: Escherichia coli uti89, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: CELL ADHESION |
 |
Structure Of Fab 297 In Complex With Influenza H1N1 A/Victoria/4897/2022 Neuraminidase
Organism: Influenza a virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: IMMUNE SYSTEM |
 |
The Secreted Adhesin Etpa Of Enterotoxigenic Escherichia Coli In Complex With The Mouse Mab 1C08
Organism: Escherichia coli etec h10407, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: CELL ADHESION Ligands: BGC |
 |
The Secreted Adhesin Etpa Of Enterotoxigenic Escherichia Coli In Complex With The Mouse Mab 1G05
Organism: Escherichia coli etec h10407, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: CELL ADHESION Ligands: BGC |
 |
Crystal Structure Of 05.Gc.W13.02 Fab In Complex With H1 Ha From A/California/04/2009(H1N1)
Organism: Influenza a virus (strain swl a/california/04/2009 h1n1), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2024-06-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, MG |
 |
Crystal Structure Of 05.Gc.W13.01 Fab In Complex With H1 Ha From A/California/04/2009(H1N1)
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-06-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of 05.Gc.W13.02 Fab In Complex With H5 Ha From A/Viet Nam/1203/2004(H5N1)
Organism: Influenza a virus (a/viet nam/1203/2004(h5n1)), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2024-06-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
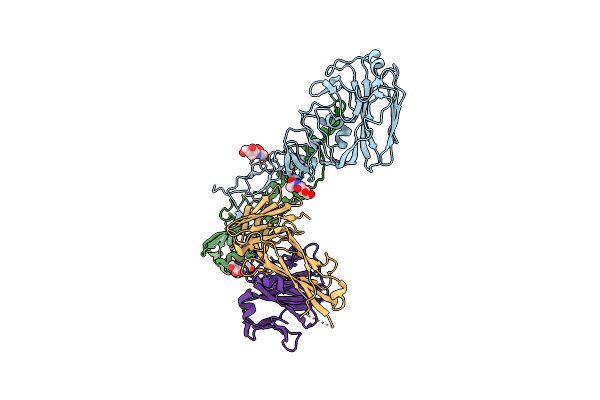 |
Crystal Structure Of 05.Gc.W2.3C10 Fab In Complex With H1 Ha From A/California/04/2009(H1N1)
Organism: Influenza a virus (strain swl a/california/04/2009 h1n1), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2024-06-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
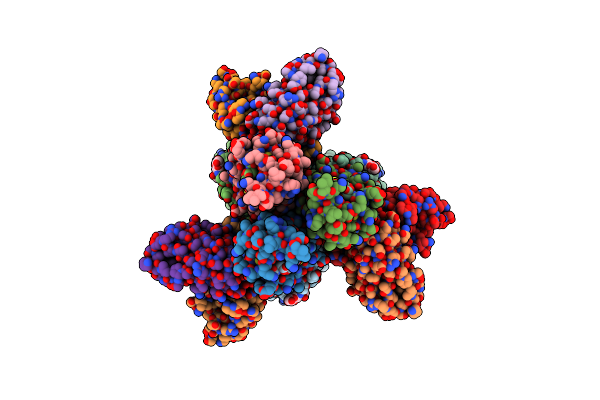 |
Cryoem Structure Of A/Solomon Islands/3/2006 H1 Ha In Complex With 05.Gc.W2.3C10-H1_Si06
Organism: Homo sapiens, Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
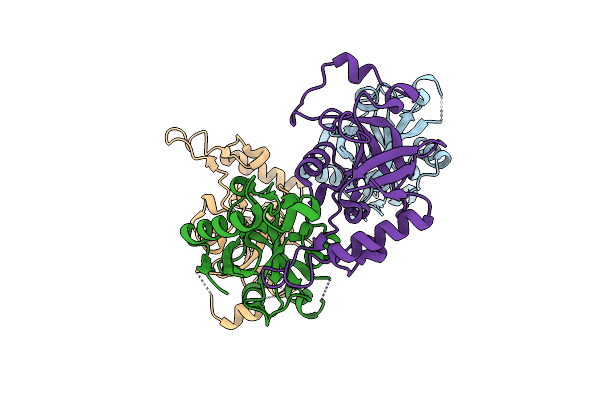 |
Organism: Wolbachia endosymbiont strain trs of brugia malayi
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-06-12 Classification: LIGASE |
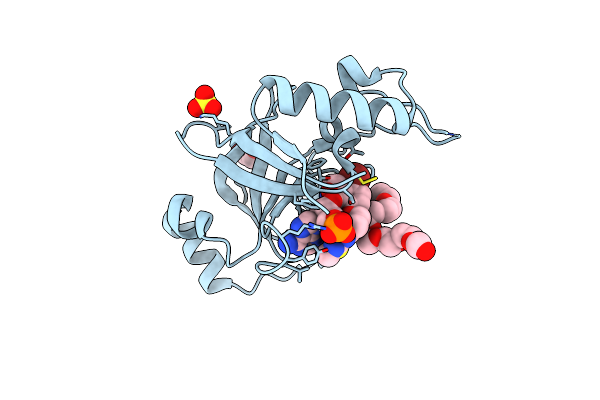 |
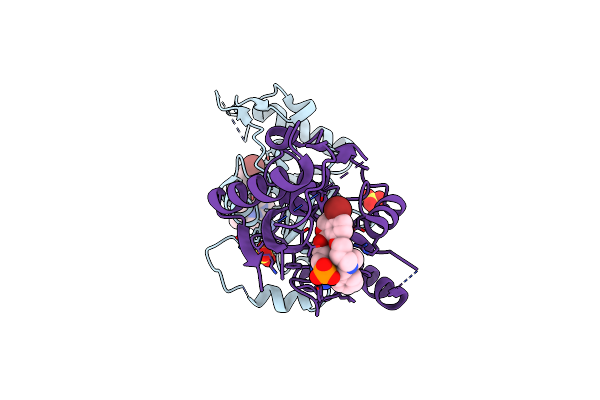
Crystal Structure Of Wolbachia Leucyl-Trna Synthetase Editing Domain Bound To Cmpd6-Amp Adduct
Organism: Wolbachia endosymbiont strain trs of brugia malayi
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-06-12 Classification: LIGASE Ligands: SO4, EYT, EDO, PE4 |
 |
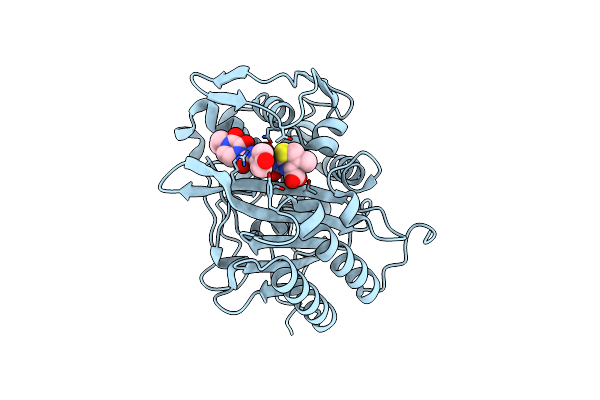
Crystal Structure Of Wolbachia Leucyl-Trna Synthetase Editing Domain Bound To Cmpd9-Amp Adduct
Organism: Wolbachia endosymbiont strain trs of brugia malayi
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-06-12 Classification: LIGASE Ligands: 7RZ, SO4 |
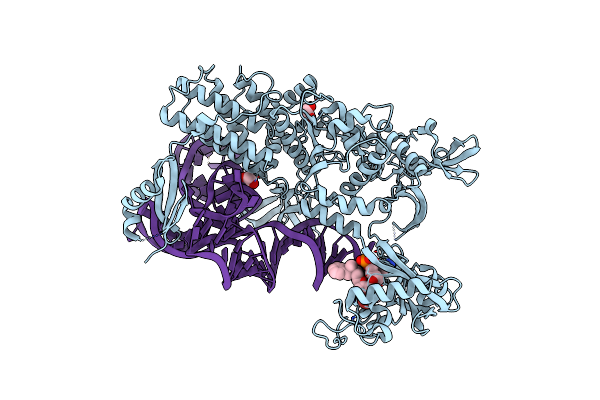 |
Ternary Complex Of E. Coli Leucyl-Trna Synthetase, Trna(Leu) And The Benzoxaborole Cmpd9 In The Editing Conformation
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-06-12 Classification: LIGASE Ligands: EDO, 7RZ |
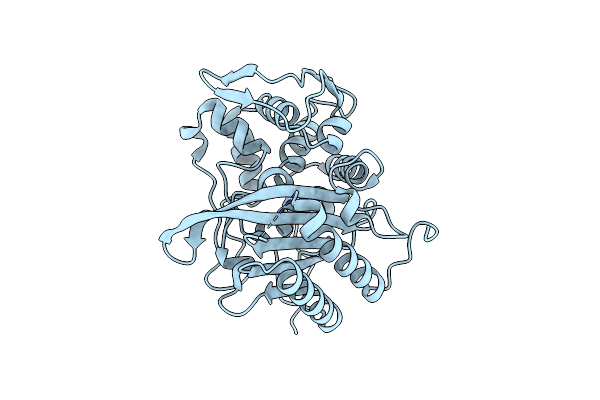 |
Crystal Structure Of The Transpeptidase Domain Of A S310A Mutant Of Pbp2 From Neisseria Gonorrhoeae Strain H041
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-03-20 Classification: HYDROLASE |
 |
Crystal Structure Of Transpeptidase Domain Of Pbp2 From Neisseria Gonorrhoeae Cephalosporin-Resistant Strain H041 In Complex With Cefoperazone
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-20 Classification: HYDROLASE Ligands: A1ADP |
 |
Crystal Structure Of Transpeptidase Domain Of Pbp2 From Neisseria Gonorrhoeae Cephalosporin-Resistant Strain H041 Acylated By Piperacillin
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-20 Classification: HYDROLASE Ligands: JPP, PEG |

